Genes, Brains, and Behavior: Imaging Genetics for Neuropsychiatric Disorders
Abstract
The majority of neuropsychiatric disorders show a strong degree of heritability, yet little is known about molecular factors involved in the pathophysiology of diseases like schizophrenia. After a brief historical introduction into the current understanding of neuropsychiatric disorders, the aim of this study is to discuss imaging genetics as a strategy to explore the pathophysiology of neuropsychiatric disorders. The candidate gene approach of imaging genetics is used for validation/replication studies of genes, whereas the hypothesis-free, noncandidate gene approach appears to be a tool for gene discovery. Besides, integration of environmental factors into neuroimaging begins to converge on neuroimaging studies of genetic variation. In the light of data from other avenues such as animal experimentation, these developments show a model of interdisciplinary research, which may lead to identifying markers for neuropsychiatric disorders.
Madness was not subject to systematic scientific scrutiny until the late 19th century, when two German-speaking psychiatrists developed diametrically opposed theories. Emil Kraepelin's opinion of madness was entirely biological. Sigmund Freud, on the other hand, formulated those as disorders of the mind principally related to early childhood experiences and psychic forces.1 Kraepelin and Freud were both born within a few hundred kilometers from each other in 1856. But they never met—neither physically nor intellectually. Although Freud accepted that some sort of mental diseases could be physiological, and in his book of 1895 entitled Project for a Scientific Psychology, he proposed a program for a scientific psychology that would be firmly based in neurophysiology,2 for decades the physiological and psychological theories of madness developed along parallel paths.1
The diagnosis of madness did of course become much more differentiated. Indeed, by the 1970s, it was known that particular biochemical pathways in the brain underpinned some psychiatric diseases such as schizophrenia.3 However, it was only in the late 1990s that the accumulation of data obtained from brain sciences led to the question of whether the whole spectrum of mental disorders could be based on a biological framework.4
Despite this, psychiatric disorders are still primarily diagnosed on clinical grounds. According to the current version of the DSM-V,5 psychiatrists have to diagnose more than 300 different manifestations of mental disorders that they believe to represent the conditions of their patients by symptoms such as hallucination, delusion, or other fairly soft criteria. One seemingly intractable problem, for example, is that a main source of information about these symptoms may not be objective because they come from the patients themselves. However, many psychiatric illnesses can be reliably detected at the level of clinical signs and symptoms according to standard reference texts—most particularly the DSM and the ICD—for diagnosis that is based mainly on symptoms, yet these classifications often lack the objective tests to draw boundaries around a particular clinical state.6–8 Therefore, the strength of the present phenomenological diagnostic guidelines derives from their reliability but not their validity.8
Since the publication of the sequence of the human genome in 2001,9 a rapid growth of different approaches to extract useful information from the genomic sequence have been evolved. These approaches included, but were not limited to, the analysis of genetic variation, gene expression, and gene products and their metabolic effects.10 As susceptibility for major psychiatric disorders show a strong degree of inheritance11 and twin studies show that heritability for various aspects of cognition, temperament, and personality ranging from 40% to 70%,12 these approaches trigger an enormous research effort to identify the biological bases of psychiatric disorders. Thus, many candidate genes have been investigated in relation to mental disorders such as schizophrenia or depression, but these investigations still remain at the theoretical level.13,14 The two very big challenges are identifying the genes and identifying the phenotypes.15 Mental disorders are not associated by a single gene. Most types of mental disorders appear to be related to multiple genes that interact with each other and with the environment and vice versa. However, it is important to mention that, although genetic heterogeneity is generally accepted to be a complicating issue for psychiatric disorders, it is not an elusive problem for gene identification.16 Moreover, there has been relatively good progress on the identification of genetic biomarkers for other multifactorial diseases triggered by complex interaction of genes and environmental factors. This is mostly evident in cancer. For example the KRAS (Kirsten rat sarcoma viral oncogene homolog) mutation in colon and lung cancers, the EGFR (epidermal growth factor receptor) mutation in lung cancer, and the BRAF (v-raf murine sarcoma viral oncogene homolog B) mutation in colon cancer have led to the identification of molecular markers to be used for diagnosis.17
Conversely, although the new data from genome-wide association studies (GWASs) sound exciting,18–22 genetic variants with a definitive effect on mental disorders such as schizophrenia phenotype are still unexplained.23,24 In addition to the complexity of genetic factors, the complexity of psychiatric phenotypes and gene–environment interactions and the lack of biologically defined diagnostic criteria are among the reasons for this problem. Of these complexities, perhaps the most discriminative feature of psychiatric disorders compared with other multifactorial disorders is the complexity of psychiatric phenotypes, the phenotype bottleneck, reflected by the lack of biologically defined diagnostic criteria, which may manifest itself in poor results of genetic findings.
The Phenotype Bottleneck and Endophenotypes
The symptoms of mental diseases are usually behavioral complications such as visual or auditory hallucinations and disorganized speech or thought that are hard to study as precise, objective, and measurable phenotypes.25 Also, some patients may exhibit a combination of such symptoms that cut across the existing disease boundaries defined by standard guidelines like the DSM. There is an overlap in genetic susceptibility across these disease boundaries. For example, schizophrenia, bipolar disorder, and schizoaffective disorders have been shown to share at least some genetic liability that cuts across the traditional binary classification of psychosis.26 Moreover, for bipolar disorder, different twin studies have shown dramatically varying concordance rates depending on the diagnostic criteria, how the samples were ascertained, and how assessments were conducted.27 Thus, one possible reason for difficulty in hunting susceptibility genes in psychiatry is that gene discovery studies have been searching for genetic effects on disease, but because disease diagnosis is not validated biologically, it does not provide a standard pool of subjects for top-down research. Nevertheless, the phenotype bottleneck is a well-known problem. Addressing this, the US National Institute of Mental Health (NIMH) has recently introduced its Research Domain Criteria Project, an alternative framework for research into psychiatric disorders.28
Also, to be helpful, psychiatric phenotypes could be divided into very small traits in different domains, like cognition or emotion, so that they are specific enough to measure reliably and quantitatively. The nature of such traits—known as endophenotypes—may be electrophysiological, biochemical, endocrinological, or anatomical, but they must be heritable. The traditional endophenotype concept was proposed for schizophrenia genetics by Gottesmann and Shields in the early 1970s to facilitate gene discovery. An endophenotype was therefore expected to reduce genetic complexity and increase genetic effect size so that the complicated effects of gene interactions and distal genotype–phenotype associations could be sufficiently narrowed to clearly identify genetic risk factors.29 Such requirements led to the establishment of certain criteria.27 It is clear that only a few, if any, endophenotypes could actually fulfill these criteria, but a handful of robust endophenotypes that have been meeting these criteria have led to the identification of genomic sequences with disease relations. One example is the use of electrophysiological measurements as an endophenotype. Deficiencies in the electrophysiological response to auditory stimulation were used to identify an association of schizophrenia with the α7 nicotinic cholinergic receptors, a transmembrane protein of neurons.30 Progressively, accumulating papers in the literature facilitate the standardization of the criteria, which increase the potential for the use of endophenotypes for psychiatric research27 to identify new genes via this reversed genetics approach.13
Birth of Imaging Genetics: Candidate Gene Approach
In parallel with these developments, advanced brain imaging techniques have led to the emergence of a so-called forward genetics approach by which, instead of discovering genes, the already known genotypic feature, a candidate gene, is intended to be correlated to an intermediate phenotype. Imaging genetics or imaging genomics are increasingly used in this context, and the endophenotypes intended to be used for this purpose are usually called an intermediate phenotype—although the latter not necessarily being different than the former by means of essential selection criteria but differ in the purpose of its use. The intermediate phenotype strategy is based on the assumption that “gene effects at the level of the brain are a more direct effect of genetic variation than is complex behavior, and will show association in carriers of risk alleles even if the carriers show no clinical diagnostic characteristics.”13
Perhaps the most attractive examples for this trend come from the earlier attempts to identify the effect of single-nucleotide polymorphisms (SNPs) on psychiatric phenotypes determined by brain imaging. Because SNPs tend to be relatively stable genetically,31 they can serve as biological markers for pinpointing a mental disease in the human genome. Use of brain imaging proves to be especially powerful for addressing this. In this way, the effect of a known genetic diversity, which has been occurring naturally among human populations, is studied by brain imaging to determine whether one form of such a variation in human genes can cause a difference in the level of such psychiatric phenotypes and hence could make people more vulnerable to mental diseases. Of genetic functions influenced by SNPs, neurobiological studies increasingly validate neural correlates that are essentially involved in brain function during health and disease. These include a wide variety of protein groups encoded by these genes with diverse functions, ranging from neural growth and survival to signal transduction or, for example, brain-derived neurotrophic factor (BDNF),32 catechol-O-methyltransferase,33 and serotonin transporter protein,34 that are associated with psychotic or mood disorders. Using a candidate gene approach, such developments in understanding the biology of cellular mechanisms encoded by these genes consequently facilitated those imaging genetics studies that aid the correlation of genetic variation by psychiatric phenotypes. The rigorous results of preliminary studies already provide a proof of principle for an understanding of psychiatric phenotypes with biological underpinnings, although recent applications of GWASs to imaging genetics studies provided conflicting results. Regarding this, one group initially studied whether a SNP in the gene encoding for BDNF has an effect on the hippocampus and determined the genotype phenotype relations as a risk factor for a particular psychiatric condition.35 It is already known that the size/morphology of the hippocampus varies among human populations and is estimated to be sufficiently heritable,36 therefore suggesting this structure as an attractive candidate for genetic correlation based on the approach of an intermediate phenotype. The hippocampus is also known for being involved in the pathogenesis of several mental disorders including schizophrenia.37–39 Reasonably, these data provide a strong basis for correlating changes seen in vivo to a potential genetic component when using a candidate gene approach. In this context, a widely expressed gene in the hippocampal formation encoding for a protein called BDNF, one of the neurotrophins that have been implicated in the neurobiology of schizophrenia, has been thought to be a good candidate. By coupling this information with an imaging genetics approach, Szeszko et al.35 reported that a certain form of the BDNF gene variation (val66met) accounted for a greater proportion of the variance in the volume of the hippocampal formation (as assessed by MRI) in patients compared with healthy volunteers. Thus, the findings led to the idea that this effect (of genetic variance of BDNF) may be greater among patients compared with healthy volunteers through hippocampal pathogenesis and/or making the hippocampus more susceptible to environmental insults.35 In contrast, recent data produced by a GWAS on hippocampal volumes failed to find the effects of BDNF val66met.40
Conversely, such controversies are not always the case. A fruitful outcome is particularly possible when functional MRI (fMRI) is used to examine the neurobiological effect of a well-validated gene. Regarding this, the first study focusing on the ε4 allele of the apolipoprotein E gene (APOE-ε4) gene published in 2000 by Bookheimer et al.41 was followed by others (Table 1). The study of Bookheimer et al., which is also the first study of imaging genetics, provides evidence that patterns of brain activation during tasks requiring memory differ depending on the APOE-ε4 allele, the genetic risk of Alzheimer’s disease. Greater brain activity is detected in the hippocampus during memory tasks among APOE-ε4 carriers compared with noncarriers (Figure 1). Thus, the authors suggested that the hyperactivity could be a compensatory mechanism used to aid task performance. Several studies42–44 have replicated these findings, whereas others have not.45 However, factors such as the utilization of different cognitive tasks or the participants’ varying age across studies appear to be influential on this discrepancy.46
| Reference | Type of association | Target gene/SNP | Target phenotype | Sample size | Related disease | Findings |
|---|---|---|---|---|---|---|
| Egan et al. 2001100 | Candidate gene-candidate phenotype | rs4680 (COMT-Val158Met) | Frontal lobe activity during during executive cognition and memory task measured by fMRI | 175 patients (SZ), 219 unaffected siblings, and 55 controls | SZ | The Val allele is associated with a reduction in performance compared with the Met allele in three cohorts. |
| Esslinger et al. 200952 | Candidate gene-candidate phenotype | rs134470 (ZNF804) | Connectivity between DLPFC and Hippocampus during memory task measured by fMRI | 115 healthy subjects | SZ, BD | GWAS supported risk variant rs1344706 impacted specifically on the connectivity between DLPFC and HF, in parallel with data found in subjects with SZ. |
| Kohli et al. 201153 | Candidate gene-candidate phenotype | rs1545843 (SLC6A15) | Hippocampal volume measured by (1H-NMR) spectroscopy MRI | 80 patients with recurrent unipolar depression and 81control subjects. | MDD | rs1545843 showed associations with alterations in hippocampal volume and neuronal integrity. |
| Flippini et al. 200954 | Candidate gene-brain-wide phenotype | rs 429358 (ApoE epsilon4) | Cortical atrophy across the brain assessed by VBM | 83 AD patients | AD | Gray matter volume decreased additively with increasing allele load in the medial and anterior temporal lobes bilaterally. |
| Braskie et al. 201155 | Candidate gene-brain-wide phenotype | rs11136000 (CLU) | White matter integrity is determined by DTI | 398 healthy subjects | AD | Risk variant was associated with lower fractional anisotropy—a widely accepted measure of white matter integrity. |
| Bis et al. 201240 | GWAS-candidate phenotype | GWAS: >3.1 million SNPs | Hippocampal volume measured by MRI | 9,232 healthy subjects | AD | The study identified two replicated hits rs17178006 and rs7294919 on chromosome 12 that correlate with hippocampal volume. |
| Shen et. al. 201068 | GWAS × brain-wide phenotype | GWAS: 530,992 SNPs | Global gray matter density determined by VBM | 733 subjects (175 AD, 354 MCI, and 204 healthy controls, HC). | MCI, AD | SNPs in the APOE and TOMM40 genes were confirmed as markers strongly associated with multiple brain regions. Other top SNPs were proximal to the EPHA4, TP63 and NXPH1 genes. |
| Hibar et al. 201170 | Multivariate GWAS × brain-wide phenotype | GWAS: 18,044 genes | 31,662 voxels of the whole brain- Structural MRI scans were analyzed by TBM | 202AD patients, 413 MCI, 237 Healthy elderly subjects | AD | Several known candidates were reidentified, as were other genes highly relevant to brain function. GAB2, which has been previously associated with late-onset AD, was identified as the top gene. |
| Ge et al. 201271 | Multivariate GWAS × brain-wide phenotype | GWAS: 448,294 SNPs, 18,043 genes | 31,662 voxels of the entire brain MRI scans analyzed by TBM | 173 AD patients,361 MCI, 206 Healthy elderly subjects | AD | GRIN2B, which encodes the N-methyl-d-aspartate glutamate receptor NR2B subunit and affects both the parietal and temporal lobes in human brains. |
TABLE 1. Examples of Imaging Genetics Studies Categorized by Candidate Gene (Tested for Either Candidate Phenotype or Brain-Wide Phenotype Association) or Noncandidate Gene (Tested for Either Candidate Phenotype or Brain-Wide Phenotype Association) Approachesa
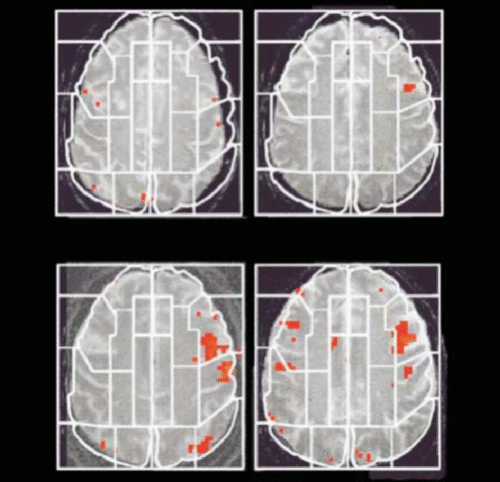
FIGURE 1. First Study of Imaging Genetics Used the Candidate Gene Approach, by Which the Brain Activation of Healthy Individuals Carrying the APOE-ε4 Allele, the Risk Variant for Alzheimer’s Disease, Is Compared With That of Noncarriers During a Memory Taska
a [A]: Examples of activation maps on single MRI planes for two carriers of the APOE-ε3 allele, which had fewer and less extensive areas of statistically significant activation (shown in red) than did [B]: two carriers of the APOE-ε4 allele. White lines indicate examples of regions of interest used for the analyses of data within subjects. (Reproduced from Bookheimer et al. 2000.41)
In one of the earlier studies of imaging genetics, an extensive work using the candidate gene approach was reported by Meyer-Lindenberg et al.47 They examined the brain activities of 126 healthy people with variants of the gene for catechol-O-metyltransferase, an important enzyme in the breakdown of neurotransmitters like dopamine. The gene is located on a region of chromosome 22 that has been linked to psychotic disorders such as bipolar disorder and schizophrenia. Dysfunction of the dorsolateral prefrontal cortex (DLPFC) has already been found using neuroimaging in schizophrenia. In this study, healthy people that carry variants of the catechol-O-metyltransferase gene associated with schizophrenia had less efficient processing in DLPFC during brain imaging, even when performance on the test was similar. This shows that disturbed processing in this region may mediate the genetic risk for schizophrenia associated with this gene by creating a vulnerable neural system that might decompensate under adverse environmental conditions. This particular function is one of many symptoms of schizophrenia, and a study showed that healthy people who can do this memory test without problems have lower activation of the DLPFC if they have the gene variant that is common among schizophrenics. They are compensating by using a different part of their brain, but it may also mean that they are at high risk of developing schizophrenia under particular environmental conditions. As suggested by Meyer-Lindenberg and Weinberger,13 this kind of study shows that genetic variance puts a person at higher risk by means of biological mechanisms converging to a specific brain circuitry: in this case, neostriatal–prefrontal and DLPFC–hippocampus circuitry that varies with the gene allele. Thus, there has been significant progress in describing the effect of common genetic polymorphisms on complex behavioral phenotypes and disease liability with the candidate gene strategy of imaging genetics. It seems like a well-replicated study of the candidate gene approach of imaging genetics might be particularly useful to address the neural system that is abnormal in a given illness.
However, the effect sizes of such studies appear to be a major source of discussion. For example, in 2002, Hariri et al.48 published a paper in Science suggesting a strong association of the short allele of serotonin system–related genetic variant 5-HTTLPR (serotonin transporter–linked polymorphic region) with activity of the amygdala measured by fMRI. However, this has been strongly criticized due to a very high effect size (28%), which appears to conflict with the recent GWAS imaging data.49
What can be deduced from this? Initial studies aimed to address the association of single neuroanatomical region with a few well-validated genetic variations in a small sample size. However, it appears that replication of initial studies of candidate genes is a must. A well-replicated study of such an approach might be useful to address the impact of genetic variation on a neural system that is abnormal in a given illness, despite the problem of false-positive findings.50 One way to address this problem might be to create a genome-wide correction of associations with imaging phenotypes with sample sizes at least in the hundreds (this would be a noncandidate gene approach, which will be discussed later). However, statistical modeling is a serious challenge in any case: one has to be careful when considering the analysis of neuroimaging data because of the statistical modeling that can produce false-positive results. This is a well-known problem of multiple comparisons and selection biases that has long been debated.51 Moreover, analysis of neuroimaging data along with genome-wide data in large populations will naturally bring additional statistical challenges.
Stronger Candidates Supported by GWAS
Nevertheless, the candidate gene (or SNP) approach now has stronger candidates supported by recent GWAS data.18–22 For example, the recently identified genome-wide significant genetic risk variants for schizophrenia21 and bipolar disorder22 naturally catalyze the efforts to explore their effects on cognition and brain structure. Reflecting this, the candidate gene approach has now gained a special momentum to find the neural correlates of genome-wide supported risk variants. Esslinger et al.52 found that the genome-wide supported risk variant for schizophrenia, rs1344706, specifically impacted on the connectivity between DLPFC and hippocampal formation, in parallel with data found in subjects with schizophrenia. In this study, 115 healthy participants were genotyped for rs1344706 and scanned by fMRI during an n-back task, related to heritable schizophrenia risk, DLPFC–hippocampal formation activity, and candidate gene variation. Task-related regional activation of DLPFC and hippocampal formation and coupling between these structures were assessed by functional connectivity analysis. Furthermore, the candidate gene approach with some convergent data from GWAS, expression studies, and animal models might be used to suggest a pathophysiological mechanism that may be accessible to drug targeting.53 Thus, the current trend of the candidate gene studies of imaging genetics is to select a GWAS-supported risk variant and test it for a candidate phenotype (intermediate phenotype). This is the typical rationale of a candidate gene–candidate phenotype association. Elaborating on this, a candidate gene can be tested for a brain-wide phenotype. This new branch of candidate gene brain-wide associations54,55 together with typical candidate gene–candidate phenotype associations is likely to be used for replication or validation of genetic effects (Table 1), although restriction of target genotypes and/or phenotypes may limit the capacity to identify important relationships.
Noncandidate Gene Approach
Another strategy of imaging genetics involve a noncandidate gene (or SNP) approach. Instead of replication/validation of a candidate gene association with a phenotype, it is a gene hunting strategy. It aims to associate either a candidate phenotype (measured by neuroimaging) with GWAS or adapts a hypothesis-free approach lacking any candidate gene or candidate phenotype: the genome-wide brain-wide association, provided by the comparison of the entire genome with the entire brain in large datasets. Table 1 provides examples for both of these strategies.
Several groups have already begun to use the former approach by which the volume of the hippocampus56 or the caudate,57 as well as activity measures of DLPFC during working memory,58 hippocampal activity during episodic memory,59 or amygdala reactivity during emotional faces task,60 has been used as structural or functional phenotypes to compare across the whole genome. Typically, each genetic variant is independently tested for its association with the phenotype, which is called a mass univariate method. Such studies initially used the structural/functional brain measures as quantitative phenotypes obtained by MRI and compared them with 300,000–500,000 SNPs in relatively smaller numbers of subjects (60–400), whereas more recent publications have used much larger scales. Four papers40,61–63 of such imaging GWASs published in Nature Genetics in 2012 demonstrate this trend. Regarding this, one study61 reported the genome-wide association meta-analyses and replication for mean bilateral hippocampal, total brain, and intracranial volumes from a large multinational consortium (N=21,151). Analysis of genome-wide SNP data and structural MRI data from healthy subjects suggests that (1) the intergenic rs7294919 variant, perhaps by regulating expression of the TESC gene encoding the protein called tescalcin, which interacts with the Na+/H+exchanger involved in the regulation of intracellular pH, cell volume, and cytoskeletal organization, was associated with decreased hippocampal volume (N=21,151); (2) rs10784502, located within HMGA2, encoding the high mobility group AT-hook 2 protein, was associated with intracranial volume (N=15,782); and (3) a suggestive association with total brain volume at rs10494373 within the DDR2 gene encoding discoidin domain-containing receptor 2 has been identified (N=6,500). Consequently, the effect of rs7294919 can now be the target of more focused studies either following the rationale of a candidate gene approach of imaging genetics, which has been published recently,64 or by complementary studies such as mapping quantitative trait loci in the mammalian syntenic region, generating mouse knockouts of nearby genes, and investigating them. As a result, with the noncandidate gene approach, a synthesis of brain imaging with GWAS data can be used to discover new genetic variants.65 However, the utility of such an approach depends on the replication of these data determined by the quality of imaging phenotypes, which is only partially achieved.66,67
Genome-Wide Analysis Matched With Brain-Wide Scan
It has been suggested that the best strategy would involve the combination of genome-wide analysis with brain-wide scanning.6 Consequently, genome-wide whole brain approaches to investigate genetic effects on neuroimaging phenotypes for identifying quantitative trait loci have been described.68–71 Technically speaking, these associations are quite challenging because of the multiple testing problem arising from possibly billions of statistical tests to be performed. The mass univariate method is frequently used for the analysis.68,69 It is based on statistics at each single voxel, focused selectively on the highest association. However, it thus eliminates the possibility of “spatially extended signals that do not win any voxel.” Moreover, because of linkage disequilibrium, variants in the same haplotype block can be highly correlated.71 Addressing this, multivariate analysis based on principal component analysis has been proposed along with other multivariate techniques including penalized and sparse regression approaches with least absolute shrinkage and selection operator.71 Whereas principal component analysis may not be useful to identify the nonlinear interactions between genetic variants and associated phenotypes, some new multivariate techniques have been reported to have better statistical power. Accordingly, Ge et al.71 reported a multivariate method to compute the association tests between 448,294 SNPs and 18,043 genes in 31,662 voxels of the entire brain across 740 elderly subjects from the Alzheimer’s disease neuroimaging initiative. Structural MRI scans were analyzed using tensor-based morphometry to compute three-dimensional maps of regional brain volume differences compared with an average template image based on healthy elderly subjects. The subsequent analysis was based on a semiparametric regression model by which the complex interaction of SNPs is modeled nonparametrically using a least-squares kernel machines approach, whereas covariate effects, such as sex and age, are modeled parametrically (i.e., linearly). A number of genes were identified as having significant associations with volumetric changes. The most associated gene was GRIN2B, encoding the N-methyl-d-aspartate glutamate receptor NR2B subunit, and it affected both the parietal and temporal lobes in human brains, reflecting the successful replication of the results of a previous study.70 Its role in Alzheimer’s disease has been widely acknowledged and studied, suggesting the validity of the approach. Figure 2 shows least-squares kernel machines results for the GRIN2B gene, which is associated with the parietal and temporal foci, both of which are known to be affected brain regions in Alzheimer’s disease. Table 1 lists some studies using mass univariate68,69 or multivariate70,71 analysis.
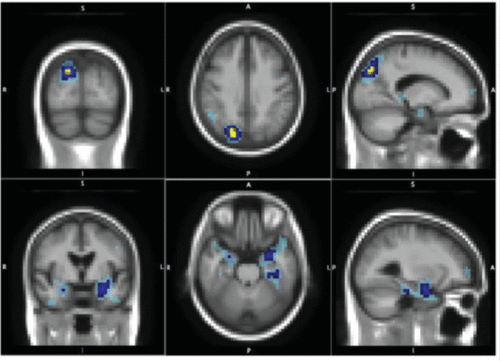
FIGURE 2. Noncandidate Gene Approach Based on Multivariate Genome-Wide Association Study × Brain-Wide Phenotype Association Identifies the GRIN2B Genea
a [Upper]: Parietal foci and [Lower]: temporal foci, both of which are known to be affected in Alzheimer's disease. Brain-wide genome-significant voxels are in yellow, brain-wide (post hoc genewise) significant voxels are in dark blue, and the light blue represents the 0.001 uncorrected significant voxels. (Reproduced from Ge et al. 2012.71)
In this context, clearly increased sample sizes require multisite studies. Thus, there are several multicenter studies conducting genome-wide association meta-analyses. For example, the Enhancing Neuroimaging Genetics through Meta-Analysis Consortium,72 which analyzes brain measures and genotypes from multiple sites across the world to improve the power to detect genetic variants that influence the brain, has pooled genome-wide genotype and neuroimaging data to replicate and identify previously unknown variants explaining a small portion of the overall heritability of certain brain structure phenotypes. The consortium analyzed the MRI scans of the brain structures of 21,000 people including healthy individuals and individuals diagnosed with depression, anxiety, Alzheimer’s disease, and schizophrenia and compared them to genetic surveys linked to relevant diseases. Regarding this, the intergenic variant rs7294919 was associated with hippocampal volume, and rs10784502, located within the gene encoding for a transcriptional regulator called high mobility group AT-hook 2, was associated with intracranial volume.61
However, the need for increased sample sizes leading to multisite studies will likely cause additional confounds due to the variability of environmental conditions. Also, the small effect sizes and the extremely high numbers of statistical comparisons in this recent trend naturally bring additional statistical challenges. Addressing these, multivariate analysis by which models for voxel-wise genome-wide associations is proposed, but it requires further examination and replication.71 The utilization of different cognitive tasks and the participants’ varying age across studies appear to be highly influential on a given neuroimaging genetic study,46 given the condition that replication is essential. In fact, replication is particularly important in the current climate, which questions the reliability of scientific findings in general73,74 and neuroimaging51,75 and imaging genetics49 in particular.
Gene–Environment Interactions
A leverage of these advances will depend on understanding the genetic influences and their interactions with the environmental context within which they operate. It is clear that there are multiple genes, signaling pathways, and interactions besides other modulatory factors such as age and sex. Moreover, it is known that environmental factors are quite influential.14 Environmental factors are relevant even for highly heritable disorders such as schizophrenia, bipolar disorder, and autism and often have higher associated risk than common genetic variants for disorders such as anxiety and depressive disorders, with the exception of eating disorders.15 Thus, it has been a major challenge to elucidate the mechanisms underlying how genes and related neural processes operate in response to environmental adversity.
Perhaps the best demonstrative series of examples come from the studies of the human serotonin transporter gene variation called the serotonin transporter–linked polymorphic region (5-HTTLPR) and stress sensitivity.76 As already supported by recent data obtained from a GWAS, an association between 5-HTTLPR and major depressive disorder has been repeatedly suggested.77 It was already reported in 1996 that this polymorphism in the promoter region of the human serotonin transporter gene (SLC6A4; also known as 5-HTT) regulates gene expression.78 Both human and animal research supports the evidence for the effect of environment on the stress sensitivity in relation to genetic variation of 5-HTT.79,80 Following this, several other studies addressed the modulatory impact of the 5-HTTLPR genotype on the effect of environmental variables.81–84 (Although some replications of these studies were not satisfactory, it is speculated that small effect size has created a statistical artifact.85) Supporting this, a moderate effect of the 5-HTTLPR genotype in relation to early life stress and levels of rumination has been shown by brain imaging studies.86 Taken together, these data represent a demonstrative example of how data from animal research, behavioral genetics, imaging genetic studies, and integrated environmental intervention are beginning to unravel the interaction of genetic variation and environmental factors. However, levels of rumination may not be a good candidate to measure the pure effect of environmental factors because resilience is likely to be affected by both genetics and environment. Moreover, the integration of environmental factors into imaging genetics in this study was obtained by the classification of subjects based on self-reports, which might threaten the validity of the research. One further step from here might be an analysis of the effect of genetic variation on the neural processing of variable psychosocial conditions. Indeed, a progress targeting this comes from the data developed by multinational teams with projects clustered in the form of European Network of Gene Environment Interactions. Among these efforts, the neuroimaging study conducted by Lederbogen et al.87 identified distinct neural mechanisms for an established environmental risk factor: urbanicity. At the first stage, participants were scanned when they were taking cognitive tests that they were told to fail. Moreover, to increase the stress level, the participants were provided with negative feedback through headphones while they were checked for stress indicators such as high blood pressure. Although there was no significant difference in the mental health between the two groups of participants, the way these two groups dealt with the stress was different, and that difference is targeted to two regions: the amygdala and the perigenual anterior cingulate cortex. These two regions were associated with increased activity for urban upbringing.87 Although this study is not devoid of its own limitations (as described in the original paper and thus will not be repeated here), it is a good example of exploring the effect of environmental risk factors (urbanicity) to analyze the neural processing of psychosocial conditions (stress). Thus, the integration of epidemiological associations to understand underlying neural mechanisms affected by environmental conditions seems feasible by the use of neuroimaging.
A further analysis of this type would be achieved by the refinement of psychosocial components in urban contexts besides to a careful consideration of the role of genetic variation. Two studies appear to fill these gaps. First, neuroimaging experiments, conducted by Zink et al. in 2008,88 have already begun to unravel how unstable social status, as an environmental risk factor, is mediated by specific neural mechanisms. The study provided a profile of the neural correlates associated with processing social hierarchies in humans. Animal studies already showed that the more subordinate position in stable social hierarchies is associated with greater stress, but in dynamic unstable social hierarchies, the dominant position is exposed to most of the stressors because of the increased competition and instability.89 Thus, to address the neural correlates of social hierarchies in humans, Zink et al. analyzed the brain activity patterns of 72 Caucasian, right-handed, healthy adults in response to different hierarchical settings (stable and unstable) during a task (e.g., multiple rounds of simple reaction time tasks or a simple visual discrimination task). The tasks allowed the players to perceive the hierarchy by the use of other players simulated in the computer screen. Interestingly, the social unstable hierarchical setting produced neural activations, which were not detected in other settings. These areas (thalamus, amygdala, and posterior cingulate) are those linked to social emotional processing and overlap with the findings of Lederbogen et al.87 Second, another stream of research with a focus on oxytocin merges with these data. Research across species has demonstrated that oxytocin plays a key role in the regulation of social cognition and behavior. As reviewed by Meyer-Lindenberg and Tost,90 it plays a crucial role in attachment, social exploration, and social recognition, as well as anxiety and stress-related behaviors. Among the relevant candidates for oxytocinergic signaling, the most extensively studied candidate is the gene coding for the oxytocin receptor.90 Among socio-behavioral phenotypes, SNPs in oxytocin receptor have been associated with positive mood,91,92 sensitivity to social support,93 deficits in mother’s sensitivity to her children’s behavior,94 and reward dependence.95 Particularly, an SNP of unknown functionality in the third intron of oxytocin receptor has emerged as an interesting candidate: rs53576 (G/A). Psychiatric relevance of this genetic variant manifests itself by studies indicating that rs53576A is overtransmitted in some families to offspring with autism spectrum disorders.88 To address the intermediate neural mechanisms of this phenomenon, Tost et al.96 used multimodal neuroimaging in a large sample of healthy human subjects to identify structural and functional alterations in oxytocin receptor risk allele carriers and their link to temperament. They found activation and interregional coupling of the amygdala during the processing of emotionally salient social cues, depending on the oxytocin receptor rs53576 genotype. In addition, they reported structural alterations in key oxytocinergic regions, which predict the lower levels of reward dependence, specifically in male risk allele carriers. Thus, in this example, accumulating studies show that oxytocin receptor gene variation (rs53576) is associated with structural and functional alterations in limbic circuitry involving the amygdala, the hypothalamus, and the cingulate gyrus, suggesting that it influences social cognition and behavior by modulating neural circuits for processing of social information and negative affect. The findings presented here converge on a previously defined circuit, the amygdala cingulate circuit, modulated by the serotonin system–related gene variant, 5-HTTLPR, which is characterized as susceptibility to environmental adversity in relation to the risk of depression,82,97,98 as already mentioned above. Taken together, these suggest that the amygdala cingulate circuit is a neural substrate, a form of neurobiological cluster, exposed to the effect of divergent molecular factors (i.e., of serotonergic and oxytocinergic systems impacting on environmental reactivity and acting as a functional unit that might play a major role for the pathophysiology of relevant psychiatric conditions). Thus, these data provide evidence for the utility of imaging genetics studies—when combined with data from different sources—in analyzing the effect of genetic variation and environmental factors on neural processing of social conditions as a susceptibility mechanism for psychiatric conditions, in this case via a mechanism dependent on the medial prefrontal cortex and amygdala.
Conclusions
Perhaps the best conclusion should start with a cancer analogy. Today, many biotech companies and start-ups already produce and sell genetic tests for molecular cancer diagnostics. Why is there no such progress in the psychiatric world? Naturally, one could argue that “cancer begins with a mutation within a single cell while diseases like schizophrenia likely involve a dysregulated neural network affected by multiple unidentified genes.” However, considering the fact that heritability of mental disorders such as schizophrenia11 is higher than many multifactorial diseases such as cancer,99 there must have been a better picture given the opportunities of genomic revolution. Despite advances in neuroscience and genetic research during the last two decades, the genetic or other biomarkers that reliably guide the diagnosis of psychiatric disorders are not satisfactory. Thus, psychiatry does not seem to benefit from the postgenomic era as desired. However, psychiatric circles need to demand it. It is a demand that can be addressed by doing things differently than before. This means a better consideration of what constitutes a phenotype. This means going beyond classical association studies by which heterogeneous patient groups selected by clinical symptoms are compared with controls. This means large-scale science. This means more interdisciplinary work by which multiple levels of analysis are achieved simultaneously. This is possible by using measures of neural activity, genetic variation, environmental context, and behavioral processes in the same experimental setup. As described above, imaging genetics have the potential to provide these opportunities. The candidate gene approach of imaging genetics can be used for validation/replication studies of genetic effects, whereas the hypothesis-free noncandidate gene approach appears to be a valuable tool for gene discovery. Large-scale research prompted by meta-analytic or multivariate imaging genomics initiatives and integration of environmental factors is another way to further the, thus far very limited, knowledge of mechanisms involved in the pathophysiology of neuropsychiatric disorders. Naturally, these developments should be considered in parallel with other large datasets. Thus, there is also a need for collecting systematic phenotype information in any given individual, at a series of different levels of environmental reactivity and resolution (molecules, cells, tissues, etc.) and then determining how these features can profitably be studied together. Hence, there is a long way to go. In this context, statistical modeling appears to be a major source of problem, along with the biases in meta-analysis and task paradigms that are conceptually linked to the genes under examination.
Nevertheless, the current progress refers to the emergence of a new period and thus holds the opportunities for the future that might lead to the identification of novel biological mechanisms underlying cognition and neuropsychiatric illness for better treatment opportunities.
1 : 150 years of Freud-Kraepelin dualism. Psychiatr Q 2007; 78:237–240Crossref, Medline, Google Scholar
2 : Project for a Scientific Psychology (1895), The Standard Edition of the Complete Psychological Works of Sigmund Freud, vol. 1. London, Hogarth Press, 1966Google Scholar
3 : Dopamine: 50 years in perspective. Trends Neurosci 2007; 30:188–193Crossref, Medline, Google Scholar
4 : A new intellectual framework for psychiatry. Am J Psychiatry 1998; 155:457–469Crossref, Medline, Google Scholar
5 : Diagnostic and Statistical Manual of Mental Disorders, 5th ed. Arlington, VA, American Psychiatric Association, 2013Crossref, Google Scholar
6 Linden DE: The challenges and promise of neuroimaging in psychiatry. Neuron 2012; 73:8–22Google Scholar
7 Tandon R: The nosology of schizophrenia: toward DSM-5 and ICD-11. Psychiatr Clin North Am 2012; 3:557–569Google Scholar
8 : Medicine. What are the right targets for psychopharmacology? Science 2003; 299:350–351Crossref, Medline, Google Scholar
9 : The sequence of the human genome. Science 2001; 291:1304–1351Crossref, Medline, Google Scholar
10 Tang J, Tan CY, Oresic M, et al: Integrating post-genomic approaches as a strategy to advance our understanding of health and disease. Genome Med. 2009; 1:35Google Scholar
11 : At issue: genes, experience, and chance in schizophrenia—positioning for the 21st century. Schizophr Bull 1997; 23:547–561Crossref, Medline, Google Scholar
12 : The genetic basis of complex human behaviors. Science 1994; 264:1733–1739Crossref, Medline, Google Scholar
13 : Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci 2006; 7:818–827Crossref, Medline, Google Scholar
14 : Genetics in Psychiatry-up-to-date review 2011. Neuroendocrinol Lett 2011; 32:389–399Medline, Google Scholar
15 : Nature and nurture in neuropsychiatric genetics: where do we stand? Dialogues Clin Neurosci 2010; 12:7–23Medline, Google Scholar
16 : Recent progress in psychiatric genetics-some hope but no hype. Hum Mol Genet 2000; 9:927–935Crossref, Medline, Google Scholar
17 : Cancer pharmacogenomics: early promise, but concerted effort needed. Science 2013; 339:1563–1566Crossref, Medline, Google Scholar
18 : Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008; 40:1053–1055Crossref, Medline, Google Scholar
19 : Psychiatric genetics gets a boost. Nat Genet 2008; 40:1042–1044Crossref, Medline, Google Scholar
20 : Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466:368–372Crossref, Medline, Google Scholar
21 : Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43:969–976Crossref, Medline, Google Scholar
22 : Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43:977–983Crossref, Medline, Google Scholar
23 : Genetics in psychiatry: are the promises met? World J Biol Psychiatry 2011; 12:81–88Crossref, Medline, Google Scholar
24 : Schizophrenia genetics: progress, at last. Curr Opin Genet Dev 2012; 22:238–244Crossref, Medline, Google Scholar
25 : Why genetic investigation of psychiatric disorders is so difficult. Curr Opin Genet Dev 2004; 14:280–286Crossref, Medline, Google Scholar
26 : The genetic deconstruction of psychosis. Schizophr Bull 2007; 33:905–911Crossref, Medline, Google Scholar
27 : Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet 2006; 22:306–313Crossref, Medline, Google Scholar
28 : DSM-5 and RDoC: progress in psychiatry research? Nat Rev Neurosci 2013; 14:810–814Crossref, Medline, Google Scholar
29 Gottesman II, Gould TD: The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 4:636–645Google Scholar
30 : Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus. Proc Natl Acad Sci USA 1997; 94:587–592Crossref, Medline, Google Scholar
31 : A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001; 409:928–933Crossref, Medline, Google Scholar
32 : Effects of BDNF polymorphisms on brain function and behavior in health and disease. Brain Res Bull 2011; 86:287–297Crossref, Medline, Google Scholar
33 : Effects of COMT polymorphisms on brain function and behavior in health and disease. Brain Res Bull 2012; 88:418–428Crossref, Medline, Google Scholar
34 : Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci 2003; 4:13–25Crossref, Medline, Google Scholar
35 : Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Mol Psychiatry 2005; 10:631–636Crossref, Medline, Google Scholar
36 : Heritability of hippocampal size in elderly twin men: equivalent influence from genes and environment. Hippocampus 2001; 11:754–762Crossref, Medline, Google Scholar
37 Jun H, Mohammed Qasim Hussaini S, Rigby MJ, et al: Functional role of adult hippocampal neurogenesis as a therapeutic strategy for mental disorders. Neural Plast 2012; 2012:854285Google Scholar
38 : Basal ganglia and limbic system pathology in schizophrenia. A morphometric study of brain volume and shrinkage. Arch Gen Psychiatry 1985; 42:784–791Crossref, Medline, Google Scholar
39 : Cognitive dysfunction in schizophrenia: unifying basic research and clinical aspects. Eur Arch Psychiatry Clin Neurosci 1999; 249(Suppl 4):69–82Crossref, Medline, Google Scholar
40 : Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet 2012; 44:545–551Crossref, Medline, Google Scholar
41 : Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med 2000; 343:450–456Crossref, Medline, Google Scholar
42 : fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer disease. Neurology 2005; 64:501–508Crossref, Medline, Google Scholar
43 : Identification of Alzheimer disease risk by functional magnetic resonance imaging. Arch Neurol 2005; 62:1881–1888Crossref, Medline, Google Scholar
44 : Increased brain activation during working memory in cognitively intact adults with the APOE epsilon4 allele. Am J Psychiatry 2006; 163:1603–1610Crossref, Medline, Google Scholar
45 : Reduced hippocampal activation during episodic encoding in middle-aged individuals at genetic risk of Alzheimer’s disease: a cross-sectional study. BMC Med 2006; 4:1Crossref, Medline, Google Scholar
46 : APOE associated hemispheric asymmetry of entorhinal cortical thickness in aging and Alzheimer’s disease. Psychiatry Res 2013; 214:212–220Crossref, Medline, Google Scholar
47 Meyer-Lindenberg A, Nichols T, Callicott JH, et al: Impact of complex genetic variation in COMT on human brain function. Mol Psychiatry 2006;11:867–877Google Scholar
48 : Serotonin transporter genetic variation and the response of the human amygdala. Science 2002; 297:400–403Crossref, Medline, Google Scholar
49 : Candidate and non-candidate genes in behavior genetics. Curr Opin Neurobiol 2013; 23:57–61. Available at doi: 10.1016/j.conb.2012.07.005Crossref, Medline, Google Scholar
50 : Imaging genetics—days of future past. Neuroimage 2010; 53:804–809Crossref, Medline, Google Scholar
51 : Voodoo and circularity errors. Neuroimage 2012; 62:945–948Crossref, Medline, Google Scholar
52 : Neural mechanisms of a genome-wide supported psychosis variant. Science 2009; 324:605Crossref, Medline, Google Scholar
53 : The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron 2011; 70:252–265Crossref, Medline, Google Scholar
54 : Anatomically-distinct genetic associations of APOE epsilon4 allele load with regional cortical atrophy in Alzheimer’s disease. Neuroimage 2009; 44:724–728Crossref, Medline, Google Scholar
55 : Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci 2011; 31:6764–6770Crossref, Medline, Google Scholar
56 : Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS ONE 2009; 4:e6501Crossref, Medline, Google Scholar
57 Stein JL, Hibar DP, Madsen SK, et al, Alzheimer’s Disease Neuroimaging Initiative Investigators: Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol Psychiatry 2011; 16:927–937Google Scholar
58 : A genome-wide survey and functional brain imaging study identify CTNNBL1 as a memory-related gene. Mol Psychiatry 2013; 18:255–263Crossref, Medline, Google Scholar
59 : Common Kibra alleles are associated with human memory performance. Science 2006; 314:475–478Crossref, Medline, Google Scholar
60 : Associations between variants near a monoaminergic pathways gene (PHOX2B) and amygdala reactivity: a genome-wide functional imaging study. Twin Res Hum Genet 2012; 15:273–285Crossref, Medline, Google Scholar
61 : Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 2012; 44:552–561Crossref, Medline, Google Scholar
62 : Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet 2012; 44:532–538Crossref, Medline, Google Scholar
63 : Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet 2012; 44:539–544Crossref, Medline, Google Scholar
64 : Functional impact of a recently identified quantitative trait locus for hippocampal volume with genome-wide support. Transcult Psychiatry 2013; 3:e287Crossref, Google Scholar
65 : Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Mol Psychiatry 2009; 14:416–428Crossref, Medline, Google Scholar
66 : Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res 2013; 47:1215–1221Crossref, Medline, Google Scholar
67 : Genetic variants affecting the neural processing of human facial expressions: evidence using a genome-wide functional imaging approach. Transcult Psychiatry 2012; 2:e143Crossref, Google Scholar
68 : Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage 2010; 53:1051–1063Crossref, Medline, Google Scholar
69 : Voxelwise genome-wide association study (vGWAS). Neuroimage 2010; 53:1160–1174Crossref, Medline, Google Scholar
70 : Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage 2011; 56:1875–1891Crossref, Medline, Google Scholar
71 : Increasing power for voxel-wise genome-wide association studies: the random field theory, least square kernel machines and fast permutation procedures. Neuroimage 2012; 63:858–873Crossref, Medline, Google Scholar
72 : The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav (Epub ahead of print, Jan 8, 2014)Crossref, Google Scholar
73 : Why most published research findings are false. PLoS Med 2005; 2:e124Crossref, Medline, Google Scholar
74 : Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA 2011; 305:2200–2210Crossref, Medline, Google Scholar
75 : A peculiar prevalence of p values just below. 05. Q J Exp Psychol (Hove) 2012; 65:2271–2279Crossref, Medline, Google Scholar
76 : Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry 2010; 167:509–527Crossref, Medline, Google Scholar
77 : Genome-wide association data provide further support for an association between 5-HTTLPR and major depressive disorder. J Affect Disord 2013; 146:438–440Crossref, Medline, Google Scholar
78 : Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 1996; 274:1527–1531Crossref, Medline, Google Scholar
79 : Serotonin transporter gene polymorphism, differential early rearing, and behavior in rhesus monkey neonates. Mol Psychiatry 2002; 7:1058–1063Crossref, Medline, Google Scholar
80 : Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 2003; 301:386–389Crossref, Medline, Google Scholar
81 : Serotonin transporter gene and childhood trauma: a G × E effect on anxiety sensitivity. Depress Anxiety 2011; 28:1048–1057Crossref, Medline, Google Scholar
82 : Neural mechanisms underlying 5-HTTLPR related sensitivity to acute stress. Am J Psychiatry 2012; 169:397–405Crossref, Medline, Google Scholar
83 : Social supports and serotonin transporter gene moderate depression in maltreated children. Proc Natl Acad Sci USA 2004; 101:17316–17321Crossref, Medline, Google Scholar
84 : The interaction of stressful life events and a serotonin transporter polymorphism in the prediction of episodes of major depression: a replication. Arch Gen Psychiatry 2005; 62:529–535Crossref, Medline, Google Scholar
85 : Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol 2006; 164:609–614Crossref, Medline, Google Scholar
86 : Neural correlates of epigenesis. Proc Natl Acad Sci USA 2006; 103:16033–16038Crossref, Medline, Google Scholar
87 : City living and urban upbringing affect neural social stress processing in humans. Nature 2011; 474:498–501Crossref, Medline, Google Scholar
88 Zink CF, Tong Y, Chen Q, et al: Know your place: neural processing of social hierarchy in humans. Neuron 2008; 58:273–283Google Scholar
89 : The influence of social hierarchy on primate health. Science 2005; 308:648–652Crossref, Medline, Google Scholar
90 : Neural mechanisms of social risk for psychiatric disorders. Nat Neurosci 2012; 15:663–668Crossref, Medline, Google Scholar
91 : Oxytocin, stress and social behavior: neurogenetics of the human oxytocin system. Curr Opin Neurobiol 2013; 23:11–16Crossref, Medline, Google Scholar
92 : Associations between the oxytocin receptor gene (OXTR) and affect, loneliness and intelligence in normal subjects. Prog Neuropsychopharmacol Biol Psychiatry 2009; 33:860–866Crossref, Medline, Google Scholar
93 : Thin-slicing study of the oxytocin receptor (OXTR) gene and the evaluation and expression of the prosocial disposition. Proc Natl Acad Sci USA 2011; 108:19189–19192Crossref, Medline, Google Scholar
94 : Common oxytocin receptor gene (OXTR) polymorphism and social support interact to reduce stress in humans. Proc Natl Acad Sci USA 2011; 108:19937–19942Crossref, Medline, Google Scholar
95 : Oxytocin receptor (OXTR) and serotonin transporter (5-HTT) genes associated with observed parenting. Soc Cogn Affect Neurosci 2008; 3:128–134Crossref, Medline, Google Scholar
96 : A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc Natl Acad Sci USA 2010; 107:13936–13941Crossref, Medline, Google Scholar
97 : The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry 2011; 68:444–454Crossref, Medline, Google Scholar
98 : Imaging genetics: perspectives from studies of genetically driven variation in serotonin function and corticolimbic affective processing. Biol Psychiatry 2006; 59:888–897Crossref, Medline, Google Scholar
99 : Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer 2002; 99:260–266Crossref, Medline, Google Scholar
100 : Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia, PNAS 2011; 98:6917–6922Crossref, Google Scholar