Cognitive Performance Related to HIV-1-Infected Monocytes
Abstract
The effect that HIV type 1 (HIV) has on neurocognition is a dynamic process whereby peripheral events are likely involved in setting the stage for clinical findings. In spite of antiretroviral therapy (ART), patients continue to be at risk for HIV-associated neurocognitive disorders (HAND), which might be related to persistence of inflammation. In a yearly assessment of HIV DNA levels in activated monocytes, increased HIV DNA copies were found in patients with persistent HAND. Furthermore, activated monocytes from patients with high HIV DNA copies secreted more inflammatory cytokines. Since these activated monocytes traffic to the CNS and enter the brain, they may contribute to an inflammatory environment in the CNS that leads to HAND.
In the era of effective antiretroviral therapy (ART), in spite of achieving suppression of plasma HIV RNA, HIV type 1 (HIV) continues to affect the central nervous system (CNS), as evidenced by persistence of HIV-associated neurocognitive disorders (HAND).1–4 Since the first report of circulating activated monocytes (CD14+CD16+) in HIV-infected individuals with neurocognitive impairment,5 this CD14+CD16+ subset has been characterized as a viral reservoir, with preferential susceptibility to infection by HIV.6–8 Activated monocytes may play an important role in HAND pathogenesis because these cells traffic to the CNS and could contribute to the inflammatory environment in the brain, leading to neurocognitive problems. We previously reported an association between HAND and high HIV DNA copies in peripheral blood mononuclear cells (PBMC) in cross-sectional ART-experienced and ART-naive cohorts.9–12 Others have suggested that activated monocytes were involved in HAND pathogenesis and are prone to an inflammatory phenotype;5,13 therefore we examined, for the first time, HIV DNA copy numbers in activated monocytes (CD14+CD16+) yearly in patients from the Hawaii Aging with HIV Cohort (HAHC). We then characterized HIV-infected activated monocytes to translate the clinical findings to ex-vivo and in-vitro models.
METHOD
Patients and Specimens
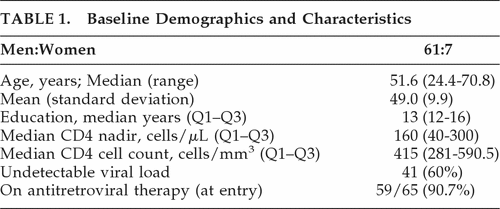
The study was approved by the University of Hawaii Institutional Review Board, with adherence to the appropriate informed-consent process. The HAHC was a longitudinal study that characterized HAND in HIV-seropositive individuals living in Hawaii from 2001 to 2006. Details of enrollment and clinical characterization were published elsewhere14,15 and are summarized in Table 1. Briefly: subjects enrolled in HAHC were HIV-infected individuals ≥50 years old and 20–40 years old, and excluded individuals with a history of head injury, learning disability, major neurologic/psychiatric disease, or brain-opportunistic disease. Baseline and annual evaluations included neurologic examination, neuropsychological testing, medical intake with demographic data, risk-behavior, HIV-1 laboratory parameters (viral load, CD4 count, and nadir CD4 count), medication histories, and comorbid illnesses. The inclusion criteria for the study were limited to participants who had an adequate number of peripheral blood mononuclear cells (PBMC; ≥20 × 106 cells per year) stored in the specimen repository for at least 3 years in order to perform the required observations.
 |
Neurocognitive Evaluations
Assessments included a macroneurologic examination, medical history questionnaire, medication/adherence history, Diagnostic and Statistical Manual of Mental Disorders–IV-based substance abuse/dependence inventory, immunologic and virologic laboratory tests, and neurocognitive testing.14,15 The neuropsychological battery included Choice and Sequential Reaction Time (CalCap; CRT/QRT), Rey Auditory-Verbal Learning Test (RAVLT), Rey-Osterreith Complex Figure (RCF) Copy and Recall, Trail-Making Tests A and B (TMA/TMB), WAIS–R Digit Symbol (SYD), Grooved Pegboard Dominant/Non-Dominant (GPD/GPN), Timed Gait (TG), Odd Man Out, fetal alcohol syndrome (FAS), Animal Naming, Boston Naming Test (BNT), and WAIS–R Digit Span (Forward and Backward). Test results were transformed to whole-number z-scores, using appropriate normative sets, as previously described.14,15 A sum score of the following tests was used to categorize degree of neuropsychological impairment: RAVLT, delayed-recall, and delayed-recognition; RCF immediate recall and copy; Odd Man Out; FAS; Digit Symbol; CalCap choice and sequential reaction times; and Grooved Pegboard dominant and non-dominant hands. Changes in neuropsychological test z-scores, grouped by cognitive domain, as noted by changes in NPZ–8 score (average of norm-adjusted z-scores for TG, GPD, GPN, TMA, TMB, SYD, CRT, and QRT), were used in the analyses.16 For the ex-vivo experiments described below, patients were categorized with research-based neurocognitive diagnoses, using the American Academy of Neurology 1991 criteria (HIV-associated dementia [HAD]; normal cognition [NC]).14,15,17
Cell-Separation and HIV DNA Assessment From the Longitudinal Cohort
Frozen PBMCs from patients who had at least 20 × 106 cells per year were separated into monocyte and non-monocyte fractions, using the Human Monocyte Enrichment Kit without CD16 Depletion (StemCell Technologies, Vancouver, BC, Canada) as per manufacturer's guidelines, which is a negative selection, leaving the CD14 cells untouched. The CD14 monocyte fractions were then separated into CD16+ (activated) and CD16− (non-activated fractions). Activated monocytes (CD14+CD16+) were labeled with the alpha CD16-biotin antibody (BioLegend; San Diego, CA) and magnetically separated by streptavidin magnetic particles (Chemicell). DNA was extracted from cell fractions and PBMC, using the QIAamp DNA Micro Extraction kit (Qiagen; Valencia, CA), with DNA quality assessed with the ND-1000 Spectrophotometer (NanoDrop Technologies; Wilmington, DE).
DNA from PBMC and PBMC subsets were assessed for HIV DNA copy numbers. Multiplex real-time PCR using HIV gag (forward 5′-GAC ATC AAG CAG CCA TGC AA-3′; reverse 5′-CTC ATC TGG CCT GGT GCA AT-3′) and β-globin (forward 5′-AGG GCC TCA CCA CCA ACT TC; reverse 5′-TCA CTA GCA ACC TCA AAC AGA CAC C-3′) primers were designed with Primer Express 3.0 software (Applied Biosystems; Foster City, CA) to amplify a 109 bp and a 111 bp PCR product, respectively. VIC-labeled HIV gag (5′ACC ATC AAT GAG GAA GCT GCA GAA TGG GA-3′) and FAM-labeled β-globin (5′-CTC CTG AGG AGA AGT CTG CCG TTA CTG CC-3′) probes were used along with standard reference plasmids with either one copy of the β-globin housekeeping gene (GenBank accession #2253431) or one copy of the HIV gag gene (GenBank accession #NC_001802) for the real-time PCR assay of HIV DNA copies. Multiplex real-time PCR master mixes were prepared with both HIV gag and β-globin primer and Taqman probe sets: 1x Taqman® Gene Expression Master Mix (Applied Biosystems; Foster City, CA), unknown, and water-to-final volume 10 μL. Plasmid dilutions from 106 to 101 gene copies were made for the standard reference curves. Non-HIV-infected Ramos β-cell DNA and water were used as negative-control DNA and HIV-infected cell DNA (OM10.1, 8E5, ACH-2; NIH AIDS Research and Reference Reagent Program; Rockville, MD) were used for positive-control DNA for inter-assay calibrations. Samples were run in triplicate on a 7900HT Real-Time PCR System (Applied Biosystems; Foster City, CA): 95°C for 10 minute; 40 cycles of 95°C for 1 minute, and 60°C for 1 minute. Analyses were completed using SDS 2.3 software (Applied Biosystems; Foster City, CA). The copy numbers of each sample gene were analyzed against the standard curves and the HIV DNA copy number per 1 × 106 cells determined.
In-Vitro and Ex-Vivo CD16+ and CD16− Monocyte Cytokines
Ex-vivo and in-vitro experiments were designed to characterize cytokine profiles of cellular subsets, which included PBMC and activated monocytes with HIV DNA.
Ex-vivo experiments were set up with specimens from three groups of patients whose PBMC were previously analyzed for HIV DNA copies as well as from HIV-negative control volunteers: 1) patients with high (log HIV DNA>2) PBMC HIV DNA copies; 2) patients with undetectable PBMC HIV DNA copies; and 3) HIV-seronegative control volunteers.9 Activated and non-activated monocytes (CD14+16+ and CD14+16−) from patients in each group were isolated as noted above and plated at a density of 3 × 106 cells per cm2 and maintained at 37°C with 5% CO2. Cells were washed 24 hours after plating to purify for adherent monocytes and maintained for 10 days in media (25% RPMI Media; Sigma, Inc., St. Louis, MO; Isocove's modified Dulvecco's medium; Irvine Scientific, Santa Ana, CA; 4 mM L-glutamine, Sigma, Inc.; 100 units/ml penicillin; 100 μg/ml streptomycin; Sigma, Inc.; 50 U/ml MCSF, Sigma, Inc.; 1 ng/ml IL3; 10% human serum; Sigma, Inc.). Equal numbers of cells were plated (96 or 48 well-plates; 9 × 105 cells/well and 3 × 106 cells/well) depending on the number of cells recovered. After 7 days, the media components were changed (excluding MCSF and IL3) and continued in culture for 3 more days. On Day 10, supernatants were obtained from CD16+ and CD16− monocyte cultures analyzed for inflammatory cytokines. Quantikine ELISA kits (R&D Systems, Inc.; Minneapolis, MN) were used according to manufacturer's instructions to assay for IL6, IL8, IL10, TNFα, sFAS, sTNFRI, sTNFRII, and MCP1. For the in-vitro experiments, monocytes were separated from commercially-available PBMC (AllCells; Emeryville, CA) using the EasySep Human CD14 Positive Selection Kit (Stem Cell Technologies; Vancouver, BC, Canada). After separation, approximately 24 × 106 monocytes were seeded in a 10-cm HydroCell Surface plate (Nunc; Rochester, NY) in 12.5 ml of Isocove's Modified DMEM (Mediatech; Manassas, VA) supplemented with 10% fetal bovine serum (Sigma-Aldrich; St. Louis, MO), 1% penicillin/streptomycin, 1% sodium pyruvate, and 1% L-glutamine. After 2 days in culture, approximately 12 × 106 monocytes were exposed to Human M-CSF (Miltenyi Biotec; Auburn, CA) at a concentration of 50 U/ml in a 6-cm Hydrocell surface plate in 8.5 ml of Isocove's Modified DMEM, as previously outlined, in order to yield activated (CD14+16+) monocytes, which were previously confirmed by flow cytometry on an aliquot of cells from the plate. After a 1-day activation period, the activated monocytes were washed and resuspended in Isocove's Modified DMEM without M-CSF. The other 12 × 106 monocytes were continued in suspension without the addition of M-CSF to yield non-activated (CD14+16−) monocytes. Each culture (in triplicate) of activated and non-activated monocytes was exposed to different strains of HIV consisting of R5– (BaL, JR–CSF), X4– (MN, LAI), and dual-tropic (p89.6) HIV with a multiplicity of infection of 0.05 in Isocove's Modified DMEM, supplemented with 2% fetal bovine serum. After an 8-hour exposure, respective viruses were washed from the cell suspensions, and the remaining viable cells were maintained in culture with Isocove's Modified DMEM, supplemented with 20% fetal bovine serum and 1% penicillin/streptomycin for a 1-day period. After recovery in DMEM supplemented with 20% FBS, both activated and non-activated monocyte cell cultures were maintained in culture for a total of 21 days post-infection with Isocove's Modified DMEM, supplemented with 10% fetal bovine serum and additional supplements as previously outlined. HIV p24 levels were assayed (ExpressBio; Westbury, NY) daily to determine optimum HIV-infected days to begin measuring cytokines. Supernatants were collected on Days 11, 15, and 19, with residual cells lysed for HIV DNA assay, had cytokines (TNFα, IL6, IL8, GMCSF, and IL1β) measured by Luminex analysis, using a 5-plex Invitrogen plate as per manufacture's guidelines. The supernatant volumes were limited; therefore, the cytokines were measured by Luminex instead of using ELISA. Supernatant cytokines from infected cell-lines were analyzed in triplicate and normalized to cytokines measured from concordant cell line cultures set up in parallel without virus.
Statistical Analyses
Pearson correlation coefficients were used to measure the associations between the NPZ8 score and each laboratory parameter at entry visit. To determine the association between HIV DNA level and NPZ8 score longitudinally, the general modeling approach included both repeated-measure models and linear mixed-effect models. One of the aims was to estimate the HIV DNA impact on NPZ8 longitudinally. To account for correlated responses for a given subject, a linear mixed-effect model was utilized to impose covariance structure of responses, to account for individual difference, and to estimate the magnitude of HIV DNA effect. In the linear mixed-effect model, NPZ8 score was regressed on log10 HIV DNA level and other covariates where unstructured covariance was assumed. Also, based on a cutoff point of 400 copies/106 cells in either PBMC HIV DNA or activated monocyte HIV DNA levels, we dichotomized our cohort into two groups (i.e., high DNA level- and low DNA level-groups) and analyzed the response profiles over time. Both repeated-measure and linear mixed-effects models were used to determine whether the shapes of the mean response profiles differed among groups. Thus, the interest was focused on whether the patterns of change in mean response were similar among groups.18–20 Cytokine measurements and analyses were performed on small numbers of specimens; therefore the data reported were descriptive.
RESULTS
Patient Demographics and Specimens
Sixty-eight patients (61 men, 7 women) were identified; their median age was 49 years. The majority of the patients' risk factors for HIV exposure were categorized as “men who had sex with men.” Of the 84% of patients on ART, 60% has undetectable HIV RNA levels at the time of entry into the cohort. The nadir CD4 cell counts ranged from 0 to 784 cells/mm3 (median: 160 cells/mm3; Q1–Q3: 40–300 cells/mm3).
Longitudinal HIV DNA Assessment
HIV DNA copies from PBMC, CD14+CD16+ (activated monocytes), CD14+CD16−, and CD14− subsets were calculated from each annual visit; and NPZ8 was also calculated from neuropsychological batteries annually. The standardized response variable, NPZ8, was assumed to be normally distributed. Pearson coefficients of correlation between NPZ8 and log copy numbers of PBMC and activated monocytes were –0.55 (p <0.001) and –0.57 (p <0.001), respectively (Figure 1[A] and Figure 1[B]). In contrast, no linear relationship with HIV DNA copies in CD14− (NS) or CD14+CD16− (NS) was found with NPZ8 score. By a linear mixed-effects model, the HIV DNA copy number in activated monocytes was strongly associated with NPZ8 (p<0.001; Figure 1[A]), but not CD4 cell count, CD4 nadir, gender, and education level. Given that HIV RNA level was fixed, a one-unit increase in log10 HIV-activated monocyte DNA copy resulted in a decrease of NPZ8 score by 0.34. A similar modeling procedure was performed for PBMC, showing that after adjusting for HIV RNA level, the HIV DNA copy number in PBMC was also strongly associated with NPZ8 score (p<0.001; Figure 1[B]); with a one-unit increase in log PBMC HIV DNA copy number resulting in a decrease of 0.32 in NPZ8 score. However, linear mixed-effect models didn't reveal significant effects of HIV copy numbers in CD14− (NS) or CD14+CD16− (p=0.09; Figure 1[C] and Figure 1[D]). Also, we validated such association between HIV DNA copy numbers and NPZ8 score by generalized estimating equation (GEE). In the GEE model, estimated effects of activated monocyte and PBMC were –0.33 (p <0.001) and –0.31 (p <0.001), respectively, which were similar to the linear mixed-effects estimates. The time effect in a linear mixed-effects model was not significant at α level 0.05. Thus, this suggested that with HIV DNA and RNA levels fixed, NPZ8 scores did not change significantly over time. To visualize this relationship of HIV DNA and NPZ8 score longitudinally, patients were divided into two groups, based on the HIV DNA copies in activated monocytes and PBMC at entry: High HIV DNA (≥400 copies/106 cells; N=32) and Low HIV DNA (<400 copies/106 cells; N=32; Figure 2[A] and Figure 2[B]). Patterns in mean NPZ8 over time seemed to be parallel between High HIV DNA and Low HIV DNA groups, with NPZ8 scores remaining stable over time (Figure 2[A] and Figure 2[B]). Both repeated-measure and linear mixed-effect models were used to test this hypothesis and confirmed that interactions between High- and Low- activated-monocytes DNA groups and time were not significant. In the linear mixed-effect model, the activated-monocytes DNA group effect was significant (p<0.0001), whereas the time effect was not (p=0.06), suggesting that mean NPZ8s were different between High- and Low-activated monocyte groups longitudinally, but no significance was observed between different time-points for a fixed HIV DNA group. The same procedure was performed by using dichotomized PBMC HIV DNA groups and showed similar results. Changes in NPZ8 over time were not statistically different for either High- or Low-PBMC groups (p=0.06), but mean NPZ8 was significantly different between PBMC DNA groups longitudinally (p<0.0001).

Activated monocyte (A), PBMC (B), non-activated monocyte (C), and lymphocyte (D) HIV DNA (log) from Years 1–5 and NPZ8 score, using a mixed-effects model to adjust for repeated measurements.
For both activated monocytes and PBMC, HIV DNA copies per million cells were strongly associated with NPZ8 scores (p <0.001) independent of baseline/nadir CD4 cell counts, gender, or education. A one-unit increase in log HIV DNA copy in activated monocytes and PBMC resulted in 0.34 and 0.32 decrease in NPZ8 score, respectively. The estimated lines assumed that HIV RNA level was 50 at baseline. No linear relationships with HIV DNA copies were found in lymphocytes (CD14−; NS) or non-activated monocytes (CD14+CD16−; NS).

Patients were divided into High (≥400 HIV DNA copies/106 cells; N=32) and Low (<400 HIV DNA copies/106 cells; N=32) and NPZ8 scores averaged over Years 1–5.
We were also interested in whether these findings could be applied to patients with undetectable HIV RNA levels. Thus, there were 32 patients who had undetectable HIV RNA levels at every annual visit. By linear mixed-effects models, we found that HIV DNA copy numbers in PBMC (coefficient: –0.2091; p<0.0001) and activated monocytes (coefficient: –0.2043; p<0.0001) were still highly associated with NPZ8 but not CD14− (NS) or CD14+CD16− (NS). Also, interaction effects between time and High/Low HIV group effect were not significant at α level 0.05 (activated-monocyte HIV groups: NS; PBMC HIV groups: NS), suggesting that mean NPZ8 profiles seemed to be parallel between High and Low HIV DNA groups. A linear mixed-effects model demonstrated that mean NPZ8s were statistically different between Low- and High-activated monocytes groups (p=0.0008) as well as Low- and High-PBMC groups (p=0.003); but no statistical significance between Low- and High-DNA groups that NPZ8 increased over time was observed for either activated monocytes or PBMCs.
Ex-Vivo and In-Vitro CD16+- and CD16−- Monocyte Cytokines
Ex-vivo cultures of activated and non-activated monocytes from patients diagnosed with HAD (N=5) and NC (N=6) and HIV-seronegative control volunteers (N=4) were established to characterize the phenotype of activated monocytes with HIV DNA isolated from HAHC patients. Activated (CD14+CD16+) and non-activated (CD14+CD16−) monocytes were placed in culture and assessed for supernatant cytokines and HIV DNA. Supernatants from activated monocytes from patients with HAD contained higher TNFα (80 [7] pg/ml), IL6 (10 [5] pg/ml), and MCP1 (998 [3] pg/ml) levels than supernatants from CD16+ monocytes isolated from NCs (TNFα: 28 [6] pg/ml; IL6: 2.8 [4] pg/ml; and MCP1: 425 [9] pg/ml) and HIV-1-seronegative controls (TNFα: 8 [2] pg/ml; IL6: 0.9 [0.1] pg/ml; and MCP1: 208 [10] pg/ml; Figure 3[A] and Figure 3[B]). Also, from the same patients with HAD, the CD16+ monocyte supernatants had higher TNFα, IL6, and MCP1 levels than the respective CD16− monocyte supernatants (Figure 3[A] and Figure 3[B]). There appeared to be no differences in CD16− monocyte TNFα, IL6, and MCP1 levels among patients with HAD, NC, or HIV-seronegative controls (Figure 3[A] and Figure 3[B]). There were also no differences in IL8, IL10, sFAS, sTNFRI, or sTNFRII levels from either CD16+ or CD16− monocytes. The patients with HAD were previously identified as having high PBMC HIV DNA7–9; and, when placed in culture, the CD14+CD16+ cells had higher HIV DNA than the CD14+CD16− cells. Because the number of specimens was small, these descriptive results provided preliminary data for the in-vitro experiments.
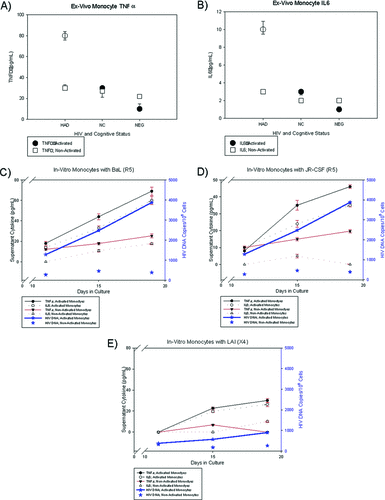
In-vivo cytokines from activated and non-activated monocytes isolated from patients diagnosed with HIV-associated dementia (HAD; N=5), normal cognition (NC; N=6), and HIV-seronegative controls (NEG; N=4); TNFα (A) and IL6 (B). Supernatants from activated monocytes from patients with HAD contained higher TNFα and IL6 than supernatants from non-activated monocytes isolated from those with NC and HIV-1-seronegative controls. In-vitro monocyte cytokine secretions showing TNFα and IL6 levels from activated and non-activated monocytes infected with R5 viruses, BaL (C) and JR-CSF (D); and X4 virus, LAI (E), on Days 11, 15, and 19. For BaL and JR-CSF infections, high cytokine levels from activated monocytes were associated with high HIV DNA copies (N=3), as compared with LAI infection (N=3). Results were descriptive because the number of specimen analyzed was small.
From the in-vitro experiments, supernatants from monocytes (activated and non-activated) infected with R5– (BaL, JR-CSF); X4– (MN, LAI); and dual-tropic (p89.6) virus were collected from Days 1–21 and assayed for HIV p24 expression, which showed infection detectable beginning on Day 11. Thus, cytokines and HIV DNA were measured on Days 11, 15, and 19. Activated monocytes infected with R5-tropic viruses (BaL and JR-CSF) had increasing TNFα (Day 19: 69.2 [2.7] and 45.7 [1.2] pg/ml, respectively) and IL6 (Day 19: 60.3 [2.1] pg/ml and 35.1 [1.1] pg/ml levels, respectively) as compared with non-activated monocytes exposed to BaL and JR-CSF (Day 19: TNFα; 25.3 [0.6] and 19.7 [0.4] pg/ml, respectively) and IL6 (Day 19: 17.8 [1.1] pg/ml and 0.0 pg/ml, respectively; Figure 3[C] and Figure 3[D]); no differences were noted in the other cytokines. The cells infected by BaL and JR-CSF on the days that cytokines were assessed also had higher HIV DNA copies in activated monocytes, as compared with non-activated monocytes (Figure 3[C] and Figure 3[D]). In contrast, X4-tropic virus (MN and LAI) cultures appeared to show no differences in TNFα or IL6 levels (nor in the other cytokines) between activated and non-activated monocytes (Figure 3[E]). Similar to the ex-vivo experiments, these results are descriptive because of the small number of samples analyzed.
DISCUSSION
The current study expands upon previous work demonstrating high HIV DNA copies in PBMC in patients with HAND.9–12 We have now identified the activated monocytes as the unique PBMC cellular subset that might be contributing to an inflammatory milieu. The data showed that patients with HAND have high HIV DNA primarily in the activated monocytes, not only at each annual visit, but patients with HAND continued to have high HIV DNA over the course of years while on ART. The results are important in understanding that patients continue to have HAND even when the HIV RNA levels remain undetectable on therapy.
In an attempt to understand mechanisms that explain the clinical findings, we then set up an ex-vivo model showing that activated monocytes isolated from patients on ART secrete more inflammatory cytokines and that the patients with HAND are those with high HIV DNA. In separate in-vitro experiments, activated monocytes infected with macrophage-tropic (CCRR5) virus, versus lympho-tropic (CXCR4) virus, produce more inflammatory cytokines. These findings support a hypothesis that activated monocytes could be a vehicle for HIV. Since monocyte trafficking, particularly, activated monocytes, to the brain has been postulated to lead to neurocognitive problems,13,21–23 the high HIV DNA levels found in activated monocytes from patients with lower NPZ8 scores is consistent with theories about HAND neuropathogenesis. The NPZ8 score has been compared with an HIV dementia scale.16 As a point of reference, an NPZ8 cutoff point was established at –2.0 standard deviations from the mean according to the AAN guidelines.17 Thus, individuals scoring below the cutoff were considered to be cognitively impaired or to have dementia, whereas scores within the cutoffs were considered to be cognitively normal.
The in-vitro and ex-vivo studies showed that activated monocytes infected with R5-tropic virus had an inflammatory cytokine profile with higher levels of TNFα and IL6. The results are similar to previous studies showing CD16 upregulation and enhanced HIV infected with MCSF; however, the present study expands upon preferential infectivity by R5-tropic virus.24–27 In circulation, the release of inflammatory cytokines could activate newly-released monocytes from the bone marrow and thus make them more prone to infection by HIV, thus setting up a chronic inflammatory state that is consistent with what is observed clinically.22,28 Our data demonstrated that activated monocytes from patients with HAND not only had higher HIV DNA copies, but produced more inflammatory cytokines than patients without HAND or HIV-1-seronegative control individuals. Others have also demonstrated that activated monocytes may play an active role, with a characteristic cytokine profile.6,7 One of the limitations of the study is that the in-vitro and ex-vivo experimental results are descriptive and require further validation to show significance.
In summary, the data showed that individuals with continued neurocognitive deficits over time are distinguished by high HIV DNA in their activated monocytes. The significance of these findings lies in the fact that, in spite of effective ART, as shown by undetectable HIV RNA levels, these patients continued to have neurocognitive deficits, with associated high HIV DNA copies in their activated monocytes. The clinical findings were consistent with the inflammatory profile that was demonstrated by activated monocytes from patients with HAND who had high HIV DNA copies. Furthermore, we showed that a similar inflammatory phenotype was found from activated monocytes that were exposed and infected by macrophage-tropic virus. The new findings, illustrating continued high HIV DNA in activated monocytes in patients with HAND, suggests a need to re-look at the current paradigm in treatment modalities. Novel therapeutic targets may need to be investigated that include monocytes and other viral reservoirs to prevent or treat HAND in patients infected with HIV.
1. : Updated research nosology for HIV-associated neurocognitive disorders. Neurology 2007; 69:1789–1799Crossref, Medline, Google Scholar
2. : Evidence for a change in AIDS dementia complex in the era of highly active antiretroviral therapy and the possibility of new forms of AIDS dementia complex. AIDS 2004; 18(Suppl 1):S75–S8Crossref, Medline, Google Scholar
3. : Human immunodeficiency virus-associated dementia: an evolving disease. J Neurovirol 2003; 9:205–221Crossref, Medline, Google Scholar
4. : HIV-associated cognitive impairment before and after the advent of combination therapy. J Neurovirol 2002; 8:136–142Crossref, Medline, Google Scholar
5. : Unique monocyte subset in patients with AIDS dementia. Lancet 1997; 349(9053):692–695Crossref, Medline, Google Scholar
6. : The contribution of monocyte infection and trafficking to viral persistence, and maintenance of the viral reservoir in HIV infection. J Leukoc Biol 2003; 74:635–641Crossref, Medline, Google Scholar
7. : The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol 2007; 178:6581–6589Crossref, Medline, Google Scholar
8. : Characteristics of activated monocyte phenotype support R5-tropic human immunodeficiency virus. Immunol Immunogenet Insights 2009; 1:15–20Crossref, Medline, Google Scholar
9. : Circulating proviral HIV DNA and HIV-associated dementia. AIDS 2005; 19:45–52Crossref, Medline, Google Scholar
10. : HIV DNA and cognition in a Thai longitudinal HAART initiation cohort: the SEARCH 001 Cohort Study. Neurology 2009; 72:992–998Crossref, Medline, Google Scholar
11. : High prevalence of human polyomavirus JC VP1 gene sequences in pediatric malignancies. Cell Mol Biol (Noisy-le-Grand) 2007; 53:4–12Medline, Google Scholar
12. : Amount of HIV DNA in peripheral blood mononuclear cells is proportional to the severity of HIV-1-associated neurocognitive disorders. J Neuropsychiatry Clin Neurosci 2009; 21:68–74Link, Google Scholar
13. : CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J Neurovirol 2001; 7:528–541Crossref, Medline, Google Scholar
14. : Higher frequency of dementia in older HIV-1 individuals: the Hawaii Aging with HIV-1 Cohort. Neurology 2004; 63:822–827Crossref, Medline, Google Scholar
15. : Cognitive impairment in older HIV-1-seropositive individuals: prevalence and potential mechanisms. AIDS 2004; 18(Suppl 1):S79–S86Crossref, Medline, Google Scholar
16. : The HIV Dementia Scale: predictive power in mild dementia and HAART. J Neurol Sci 2007; 260(1–2):11–15Crossref, Medline, Google Scholar
17.
18. : Applied Longitudinal Analysis. Hoboken, NJ, Wiley, 2004Google Scholar
19. : Linear Mixed Models for Longitudinal Data. New York, Springer, 2009Google Scholar
20. : SAS for Mixed Models. Cary, NC, SAS, 2006Google Scholar
21. : Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and non-human primate studies. J Neurovirol 2008; 14:318–326Crossref, Medline, Google Scholar
22. : Monocyte/macrophage traffic in HIV and SIV encephalitis. J Leukoc Biol 2003; 74:650–656Crossref, Medline, Google Scholar
23. : Invasive chronic inflammatory monocyte phenotype in subjects with high HIV-1 viral load. J Neuroimmunol 2004; 157(1–2):93–98Crossref, Medline, Google Scholar
24. : Endogenous macrophage CSF production is associated with viral replication in HIV-1-infected human monocyte-derived macrophages. J Immunol 1995; 154:5528–5535Medline, Google Scholar
25. : Effects of rhM-CSF expressed in silkworm on cytokine productions and membrane molecule expressions of human monocytes Acta Pharmacol Sin 2000; 21:797–801Medline, Google Scholar
26. : Enhanced HIV replication in macrophage colony-stimulating factor-treated monocytes. J Immunol 1991; 146:298–306Medline, Google Scholar
27. : Macrophage colony-stimulating factor antagonists inhibit replication of HIV-1 in human macrophages. J Immunol 2000; 164:4955–4960Crossref, Medline, Google Scholar
28. : Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infections. Proc Natl Acad Sci U S A 1989; 86:675–679Crossref, Medline, Google Scholar