Proteomic Analysis of Corticobasal Degeneration: A Case Study of Corticobasal Degeneration at the Proteome Level
Abstract
Corticobasal degeneration (CBD) is an adult-onset progressive neurodegenerative disorder. However, its pathogenic mechanisms underlying this disorder remain poorly understood. The authors examined changes of proteome profiles between three nondemented comparison brains and a CBD brain based on two-dimensional gel electrophoresis. Two up-regulated proteins in the CBD brain were identified as protein-L-isoaspartate (D-aspartate) O-methyltransferase and cofilin 1 (non-muscle), and six down-regulated proteins were identified as carbonyl reductase [NADPH] 1, two of peptidyl-prolyl cis-trans isomerase A, ubiquitin carboxyl-terminal hydrolase isozyme L1, phosphoglycerate mutase 1 (brain) and chain A of human peroxiredoxin 5. Subsequently, the possible relevance of these changes was analyzed.
Corticobasal degeneration is a progressive neurological disorder characterized by nerve cell loss and atrophy (shrinkage) of multiple areas of the brain including the cerebral cortex and the basal ganglia. Symptoms, which typically begin at or around age 60, are similar to those found in Parkinson disease. These symptoms include poor coordination, akinesia (an absence of movements), rigidity (a resistance to imposed movement), disequilibrium (impaired balance); and limb dystonia (abnormal muscle postures). Other symptoms such as cognitive and visual-spatial impairments, apraxia (loss of the ability to make familiar, purposeful movements), hesitant and halting speech, myoclonus (muscular jerks), and dysphagia (difficulty swallowing) may also occur.
Corticobasal degeneration usually progresses slowly over the course of 6 to 8 years. Death is generally caused by pneumonia or other complications of severe debility such as sepsis or pulmonary embolism.
To date, there is no treatment available to slow the course of corticobasal degeneration, and the symptoms of the disease are generally resistant to therapy. Drugs used to treat Parkinson disease-type symptoms do not produce any significant or sustained improvement. Clinical features, electrophysiologic tests, autonomic tests, neuropsychologic tests and imaging technology have been demonstrated useful in the diagnosis of CBD.1–6
The term proteome refers to a specific PROTEin complement expressed by a given genOME. Proteomics, the study of the proteome, is an ideal technology for detecting changes in proteins profiling as it allows comparison of two or more samples on a relatively global level.7 Studies of postmortem human brains have become an increasingly essential component of the effort to understand the neurobiology of neurodegenerative disorders, particularly in light of the new opportunities to apply the proteomics approaches.8
Experiments were carried out under the assumption that changes in specific functional proteins play a key role in the pathogenesis of CBD. To gain insight into the pathogenic mechanisms of CBD, and to identify biomarker candidates, we employed two-dimensional gel electrophoresis (2-DE) to compare a case of postmortem human brain with CBD and three age-matched, nondemented comparison brains. This analysis facilitated observation of changes within the proteome of the CBD brain measured against nondemented comparison group.
The core technologies of proteomics are 2-DE and mass spectrometry. 2-DE is an effective method for identifying qualitative and quantitative differences between proteomes expressed in various tissues.9 Currently several challenges exist for 2-DE including overall displayed protein spot number, isoelectric focusing of very basic proteins, electrophoresis of very high-molecular weight proteins, and hydrophobic proteins, as well as detection of low abundance proteins. Although the number of proteins displayed by 2-DE is much less than the estimated 50,000–100,000 proteins expressed in the human brain, 2-DE in its current form enables the separation of thousands of the individual proteins that constitute a large portion of a given tissue’s proteome. In our study, ten gel spots of significantly different intensities between the CBD brain and contols were found, the proteins within them were subsequently identified by in-gel trypsin digestion followed by mass spectral analysis or N-terminal sequencing.
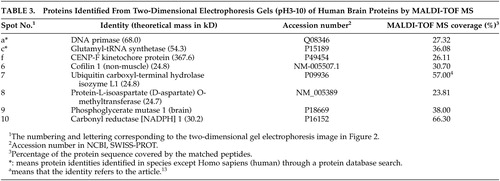
CBD is associated with neuronal and glial hyperphosphorylated tau deposits. Strong expression of the phosphorylated form of glycogen synthase kinase-3beta (GSK-3beta) was found in dystrophic neuritis of CBD, which contributes to irregular tau phosphorylation.10 We detected ten proteins that were differently expressed between CBD and nondemented comparison group and proceeded to identify eight of them as peptidyl-prolyl cis-trans isomerase A, cofilin 1 (non-muscle), chain A of human peroxiredoxin 5, ubiquitin carboxyl-terminal hydrolase isozyme L1, protein-L-isoaspartate (D-aspartate) O-methyltransferase, phosphoglycerate mutase 1 and carbonyl reductase [NADPH] 1. Nine unchanged proteins are also identified, which will enrich a 2-DE gel database of human brain proteins.
MATERIALS AND METHODS
Materials
Immobilized pH-gradient (IPG) strips were purchased from Amersham Pharmacia Biotech, Sweden. Dithiothreitol (DTT), acrylamide, N, N’-methylenebisacrylamide, CHAPS, TEMED, TPCK-treated trypsin and protease inhibitors were obtained from Sigma, USA. Trifluoroacetic acid (TFA) was purchased from Aldrich and acetonitrile (ACN) was from Fisher (New Jersey, USA).
Human Brain Tissues
The proteins of four postmortem human brains, including white and gray matter, were analyzed in this study. These temporal and parietal lobes were obtained from hospital 301 in Beijing. The brains were from four individuals: 1) a 73-year-old Chinese male and 2) a 90-year-old Chinese male who died of multiple system organ failure; 3) an 84-year-old Chinese male, dead of respiratory failure and a functional nervous disorder characterized by motor disturbances, and 4) an 88-year-old Chinese male, dead of CBD. This CBD brain was identified by paraffin-embedded sections cut and stained with HE, Bielschowsky, Holzer, Bodian, LFB and Gallyas/tau for light microscopy. The brain tissues were removed during autopsy within the time of 24h postmortem, and stored at minus 70? until further processing. All four postmortem human brains were stored at minus 70°C for less than a month before experiments were performed.
Preparation of Protein Samples and 2-DE
Whole temporal or parietal lobes were suspended and homogenized in 40ml of 0.2 mol/L NH4HCO3 containing protease inhibitors at 4? using a homogenizer. Then these samples were centrifuged at 19000×g for 1h. The supernatant was combined with three times volume of cooled ethanol. After stored at minus 20? overnight, the protein pellets were obtained by centrifugation and then lyophilized. 2-DE was subsequently performed as reported.11,12
N-terminal Sequencing and Similarity Search
After electro-transfer onto a PVDF membrane, the selected stained protein spots were excised and transferred into the cartridge of a ProciseTM 494cLC sequencer (Applied Biosystems, Foster City, CA, USA). The N-terminal amino acid sequence obtained was searched for sequence similarity against SWISS-PROT on the conventional program FASTA.
Sample Preparation and Mass Spectrometry Analyses
The protein spots of interest were excised from the Coomassie Brilliant Blue-staining gel and then trypsin digested as reported.11 For ESI-MS, the digests were added with 50μl 0.1% TFA, 1% ACN aqueous solution. The solution was centrifuged and 20μl of supernatant was applied for the analysis of ESI-MS. For MALDI-TOF MS, the digests were added with 15μl 0.1% TFA aqueous solution. The solution was centrifuged and 1μl of supernatant was applied for the analysis of MALDI-TOF MS.
RESULTS
2-DE of human temporal and parietal lobe proteins
Two-dimensional gel electrophoresis maps were constructed for human temporal and parietal lobe proteins. To investigate more proteins in a wider pH range, we made separate maps for the wide pH range (3 to 10). The second-dimension of electrophoresis was performed with a 12.5% SDS-PAGE. 2-DE maps of human nondemented comparison and CBD brains are shown in Fig. 1 A-B. For each sample over 1000 protein spots were resolved in a 2-DE gel (16×18 cm) with silver-staining.
Significant Changes in Proteins Between Human CBD Brain and Nondemented Comparison Group
Twelve independent, paired analytical 2-DE gels of human temporal or parietal lobes proteins were run and subsequently silver-stained: 2-DE runs of 88-year human CBD and 90-year nondemented comparison temporals proteome repeated in triplicate, 2-DE runs of 88-year human CBD and an 84-year nondemented comparison parietal or temporal proteome repeated four times (once for parietal and three times for temporal), 2-DE runs of 88-year human CBD and 73-year nondemented comparison temporal or parietal proteome repeated five times (once for parietal and four times for temporal).
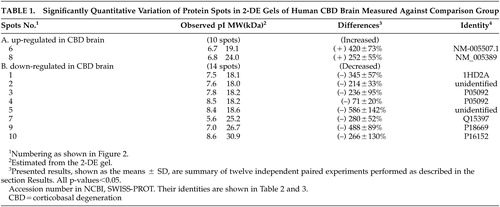
With the help of the software ImageMaster 2D v3.10, protein spots in 2-DE gels were compared between human CBD and nondemented comparison brains in these twelve independent experiments. Upon the summation of experimental results, distinct variations were identified specifically as those with a significance value of p<0.05 and were subsequently classified into the following two categories shown in Table 1 depending on observed protein levels: (A) up-regulated in human CBD brain, (B) down-regulated in human CBD brain. Ten spots were chosen for the above two categories. Figure 1 C-H displays zoomed in images of all significantly changed protein spots that appear in Figures 1 A and B. Each of protein spots that were selected as changed in this paper has a consistent increase or decrease in all twelve experiments, and the ranges of changes are all larger than 50%. These spots’ positions were marked with numbers in the 2-DE map in Figure 2; the estimated pIs, estimated molecular masses, a percent difference and identities of varied spots are summarized in Table 1.
Identification of Changed and Unchanged Proteins Between the CBD Brain and Nondemented Comparisons
Eight changed protein spots and nine unchanged spots between human CBD brain and nondemented comparisons were analyzed and identified with N-terminal Edman sequencing, LC-ESI-MS or MALDI-TOF MS. Two up-regulated protein spots in human CBD brain were identified as protein-L-isoaspartate (D-aspartate) O-methyltransferase and cofilin 1 (non-muscle). Six down-regulated protein spots in human CBD brain, were identified as carbonyl reductase [NADPH] 1, two of peptidyl-prolyl cis-trans isomerase A, ubiquitin carboxyl-terminal hydrolase isozyme L1, phosphoglycerate mutase 1 (brain) and chain A of human peroxiredoxin 5. Information about these identified protein spots, which are lettered or numbered as shown in the 2-DE map in Figure 2, is summarized in Tables 2 and 3.
DISCUSSION
We have found that a CBD brain shows a pattern of increased and decreased proteins, specifically two increased and eight decreased proteins. The protein differences observed in the CBD brain are unlikely to be caused by experimental artifacts. This can be justified by the following: each of protein spots selected as changed level in this paper, has a consistent increase or decrease in all twelve independent paired experiments, the range of changes are all larger than 50%, recent comparative proteome analysis of different-aged postmortem brains using equivalent methods in our laboratory revealed significant protein differences between human young and old brains.12 In this study we found increased levels of protein-L-isoaspartate (D-aspartate) O-methyltransferase and cofilin 1 (non-muscle), and decreased levels of carbonyl reductase [NADPH] 1, peptidyl-prolyl cis-trans isomerase A, ubiquitin carboxyl-terminal hydrolase isozyme L1, phosphoglycerate mutase 1 (brain) and chain A of human peroxiredoxin 5 in the CBD brain compared with nondemented comparisons.
Protein-L-isoaspartate (D-aspartate) O-methyltransferase (PIMT) has been shown to initiate the production of abnormal L-isoaspartyl residues,14 animal models of PIMT knockout have demonstrated a reduction of life span15 and genetic pimt knockout mice showed significant growth retardation and succumbed to fatal seizures at an average of 42 days after birth.16 Furthermore, increased levels of L-isoaspartyl residues in plasma proteins were measured in uremic patients.17 Our findings of up-regulated PIMT in the CBD brain gives us that PIMT may have the potential to be developed for human therapeutic use in repairing damaged proteins in diseases.
Cofilins are essential regulators of actin filament turnover.18 Up-regulated cofilin-1 in CBD suggests an acceleration in actin filament depolymerization.
Carbonyl reductases are NADPH-dependent, mostly monomeric, cytosolic enzymes with broad substrate specificity for many endogenous and xenobiotic carbonyl compounds.19 Competitive cDNA library screening and PCR analysis have been used to examine gene expression in mouse lung adenocarcinomas. Carbonyl reductase was found to be under-expressed in lung adenocarcinomas compared with normal lungs.20 Moreover, carbonyl reductase is down-regulated in peripheral tissues during fever in rats.21 Umemoto et al.22 showed that decreased carbonyl reductase expression in human epithelial ovarian cancer is associated with retroperitoneal lymph node (RLN) metastasis and poor survival. Our results of down-regulated carbonyl reductase [NADPH] 1 in the CBD brain provides further evidences that decreased level of carbonyl reductase is related with diseases.
Two different protein spots, number 3 and number 4, were both identified as peptidyl-prolyl cis-trans isomerase A (PPIase). Interestingly, the quantity of PPIase represented in spot number 4 was decreased by (–) 71% in the CBD comparing with nondemented brains, while the quantity of PPIase represented in spot number 3 was decreased by (–) 236% in the CBD brain. From the 2-DE gel image, protein spots of number 3 and number 4 appear to have similar molecular weights, while the observed pI of number 3 and number 4 is 7.8 and 8.3, respectively. These results most likely suggest that protein spot number 3 is the phosphorylated form of PPIase and protein spot number 4 is the unphosphorylated form. PPIases accelerate the folding of proteins and catalyze the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. PPIases were recently also found to be down-regulated in Alzheimer’s disease.9 Our results demonstrate that PPIase levels are related with CBD suggesting a possible link between PPIase and dementias.
Ubiquitin-protein hydrolase L1 (UCHL1) is involved both in the processing of ubiquitin precursors and of ubiquinated proteins. This enzyme is a thiol protease that recognizes and hydrolyzes a peptide bond at the C-terminal glycine of ubiquitin. Leroy et al identified, in a German family with Parkinson’s disease, a missense mutation in the UCHL1 gene. He went on to demonstrate that this mutation, Ile93Met, causes a partial loss of the catalytic activity of this thiol protease, which could lead to aberrations in the proteolytic pathway and aggregation of proteins.23 We showed that ubiquitin carboxyl-terminal hydrolase isozyme L1 is apparently downregulated in the CBD brain.
The interconversion of 3-phosphoglycerate and 2-phosphoglycerate during glycolysis and gluconeogenesis is catalyzed by phosphoglycerate mutase. Our findings that phosphoglycerate mutase 1 was down-regulated in CBD leads one to invision that impaired energy metabolism plays a role in the development of CBD.
As an antioxidant enzyme, chain A of human peroxiredoxin 5 reduces hydrogen peroxide and alkyl hydroperoxide with reducing equivalents provided through the thioredoxin system and is involved in intracellular redox signaling. The result that chain A of human peroxiredoxin 5 was down-regulated in CBD suggests that oxidants may play a role in CBD.
With the arrival of effective symptomatic treatments and the promise of drugs that may delay progression, the critical task now becomes diagnosis of CBD at an early stage of the disease. To diagnose CBD earlier and more accurately, attention has been directed toward peripheral biomarkers. In combination with readily available compound libraries produced by combinatorial chemistry and advances in high-throughput screening technologies, proteomics has led to an increased number of potential new lead compounds.24 Some of these above mentioned proteins that demonstrated significant changes in protein level could possibly serve as diagnostic or prognostic biomarkers of CBD at an early stage, and as potential new drug targets for therapeutic use in CBD through further research.
Investigation of the postmortem human brain is an essential component of the current effort to unravel psychiatric disorders. Increasing the number of postmortem brain specimens available for study is central to the success of this arm of the scientific assault on psychiatric illness.8 In this study we employed a case of postmortem CBD brain and three nondemented comparisons. Our results are preliminary and more cases of postmortem CBD brains are needed to confirm or modify these results.
ACKNOWLEDGMENTS
The authors thank Professor Luning Wang from the 301 Hospital in Beijing, China for the professional postmortem diagnoses and providing postmortem human brain tissues.
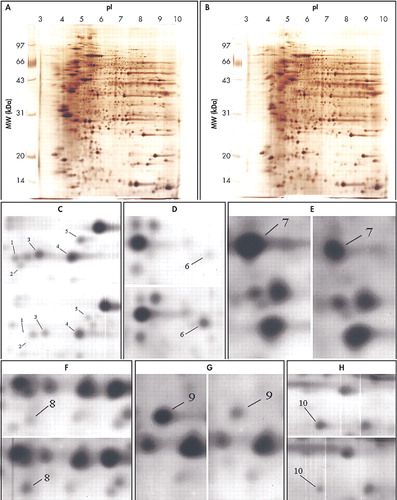
FIGURE 1. Silver-Stained Two-Dimensional Gel Electrophoresis Maps of Temporal Lobes From Human Nondemented Comparison and CBD Brains
A. 2-DE image of the temporal lobe’s proteins from a nondemented comparison brain of 90-year old.
B. 2-DE image of the temporal lobe’s proteins from a CBD brain of 88-year old.
(C), (D), (E), (F), (G), (H) All expression-changed protein spots in the 2-DE gels, which differed significantly in the expression level between CBD and nondemented brains, were zoomed in. The left or top maps represent the nondemented brain, the right or bottom representing the CBD brain. The numbers refer to Figure 2.
CBD=corticobasal degeneration
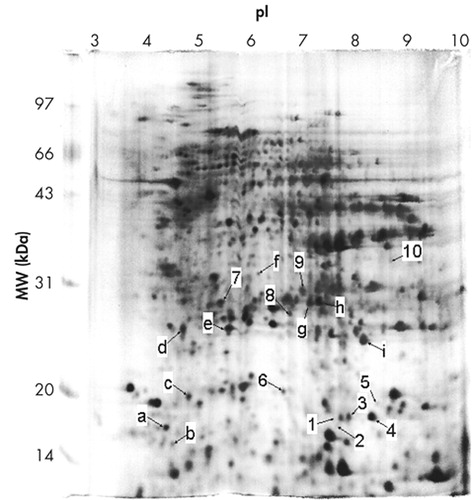
FIGURE 2. Indication of Identified Protein Spots and Variably Expressed Protein Spots Between Human Nondemented Comparison and CBD Brains
Numbering as noted in Table 1
Lettering as noted in Table 2 and Table 3
CBD=corticobasal degeneration
 |
 |
 |
1 Kashmere J, Camicioli R, Martin W: Parkinsonian syndromes and differential diagnosis. Curr Opin Neurol 2002; 86 (3):501–519Google Scholar
2 Zhang L, Murata Y, Ishida R, et al: Differentiating between progressive supranuclear palsy and corticobasal degeneration by brain perfusion SPET. Nucl Med Commun 2001; 22 (7):767–772Google Scholar
3 Charpentier P, Kreisler A, Defebvre L, et al: Differential diagnosis between corticobasal degeneration and Parkinson's disease assessed by HMPAO SPECT. Eur J Nucl Med 2001; 28 (8):PS59Google Scholar
4 Trojanowski JQ, Dickson D: Update on the neuropathological diagnosis of Frontotemporal Dementias. J Neuropathol Exp Neurol 2001; 60 (12):1123–1126Google Scholar
5 Hosaka K, Ishii K, Sakamoto S, et al: Voxel-based comparison of regional cerebral glucose metabolism between PSP and corticobasal degeneration. J Neurol Sci 2002; 199 (1–2):67–71Google Scholar
6 Moretti R, Torre P, Antonello RM, et al: Comolex distal movement in cortical-basal ganglionic degeneration. A functional evaluation. Funct Neurol 2002; 17 (2):71–76Google Scholar
7 Skynner HA, Rosahi TW, Knowles MR, et al: Alterations of stress related proteins in genetically altered mics revealed by two-diemensional differential in-gel electrophoresis analysis. Proteomics 2002; 2:1018–1025Crossref, Medline, Google Scholar
8 Lewis DA: The human brain revisited: opportunities and challenges in postmortem studies of psychiatric disorders. Neuropsychopharmacology 2002; 26 (2):143–154Google Scholar
9 Schonberger SJ, Edgar PF, Kydd R, et al: Proteomic analysis of the brain in Alzheimer’s desease: Molecular phenotype of a complex disease process. Proteomics 2001; 1:1519–1528Crossref, Medline, Google Scholar
10 Ferrer I, Barrachina M, Puig B: Glycogen synthase kinase-3 is associated with neuronal and glial hyperphosphorylated tau deposits in Alzheimer's disease, Pick's disease, progressive supranuclear palsy and corticobasal degeneration. Acta Neuropathol (Berl) 2002; 104(6):583–591Medline, Google Scholar
11 Chen W, Ji J, Zhao R, et al: Comparative proteome analysis of human temporal cortex lobes by two-dimensional electrophoresis and identification of selected common proteins. Neurochem Res; 2002. 27 (9): 871–881Google Scholar
12 Chen W, Ji J, Xu X, et al: Proteomic Comparison Between Human Young and Old Brains by Two-dimensional Gel Electrophoresis and Identification of Proteins. Int J Devl Neuroscience 2003; 21 (4):209–216Google Scholar
13 Langen H, Berndt P, Röder D, et al: Two-dimensional map of human brain proteins. Electrophoresis 1999; 20: 907–916Google Scholar
14 Thapar N, Griffith SC, Yeates TO, et al: Protein repair methyltransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Unusual methyl-accepting affinity for D-aspartyl and N-succinyl-containing peptides. J Biol Chem 2002; 277(2): 1058–65Google Scholar
15 Young AL, Carter WG, Doyle HA, et al: Structural integrity of histone H2B in vivo requires the activity of protein L-isoaspartate O-methyltransferase, a putative protein repair enzyme. J Biol Chem 2001; 276(40):37161–5Crossref, Medline, Google Scholar
16 Kim E, Lowenson JD, MacLaren DC, et al: Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc Natl Acad Sci U S A 1997; 94(12):6132–7Crossref, Medline, Google Scholar
17 Perna AF, Castaldo P, De Santo NG, et al: Plasma proteins containing damaged L-isoaspartyl residues are increased in uremia: implications for mechanism. Kidney Int 2001; 59(6):2299–308Crossref, Medline, Google Scholar
18 Vartiainen MK, Mustonen T, Mattila PK, et al: The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell 2002; 13(1): 183–94Google Scholar
19 Forrest GL, Gonzalez B. 2000. Carbonyl reductase. Chem Biol Interact 129(1–2):21–40Google Scholar
20 Lin L, Wang Y, Bergman G, et al: Detection of differentially expressed genes in mouse lung adenocarcinomas. Exp Lung Res 2001; 27(3):217–29Crossref, Medline, Google Scholar
21 Ivanov AI, Scheck AC, Romanovsky AA: Expression of genes controlling transport and catabolism of prostaglandin E2 in lipopolysaccharide fever. Am J Physiol Regul Integr Comp Physiol 2003; 284:R698-R706Google Scholar
22 Umemoto M, Yokoyama Y, Sato S, et al: Carbonyl reductase as a significant predictor of survival and lymph node metastasis in epithelial ovarian cancer. Br J Cancer 2001; 85(7):1032–6Crossref, Medline, Google Scholar
23 Leroy E, Boyer R, Auburger G: The ubiquitin pathway in Parkinson’s disease. Nature 1998; 395:451–452Crossref, Medline, Google Scholar
24 Steiner S, Witzmann FA: Proteomics: Applications and opportunities in preclinical drug development. Electrophoresis 2000; 21:2099–2104Crossref, Medline, Google Scholar