Prevalence of Alzheimer's Disease and Apolipoprotein E Allele Frequencies in the Old Order Amish
Abstract
The authors examined the prevalence of Alzheimer's disease and apolipoprotein E allele frequencies in the Old Order Amish. A lower frequency of dementia in the Amish does not appear to be due to a reduced E4 frequency.
Alzheimer's disease (AD) appears in some families to be transmitted as an autosomal dominant disorder, and in families with AD with early age of onset, three genes may account for the majority of affected individuals.1–3 In families with later age of onset, no single genetic locus has been identified, although alleles of the apolipoprotein E (APOE) locus may influence risk of developing the disease. There are three APOE alleles: E2, E3, and E4; both familial and sporadic AD patients have a higher frequency of the E4 allele than the general population,4 whereas the E2 allele may have a protective effect.5
The search for other genes that influence risk for developing AD has led to the study of populations with an apparent altered risk and genetic background, such as the Volga Germans6 and, recently, the Amish of Indiana and southern Michigan.7 The Amish are a valuable resource for genetic studies because of their large family sizes and well-defined ancestry. Pericak-Vance et al.7 have recently found a lower frequency of dementia in the Amish as well as a lower frequency of the E4 allele, and they hypothesize that these two findings may be related.
We describe a study with a different Amish population, the Old Order Amish of Lancaster County, Pennsylvania, that does not support the hypothesis that a lower frequency of AD in the Amish may be accounted for by a reduced prevalence of the E4 allele.
METHODS
Genetic studies of the Amish at Johns Hopkins have identified 40 three-generation pedigrees (the “Forty Families”), from which cell lines have been made on all individuals (n=124, ages 41–90). The Amish community is organized into a series of church districts. The Amish socialize frequently within and between these districts, and they gather every 2 weeks for church services, as active membership in the community is an important principle of Amish belief. Over a 6-month period, a trained Amish research assistant was able to review the behavior of all individuals in three contiguous church districts and of all the members of the Forty Families. Any individuals she suspected to be affected by dementia were examined by a psychiatrist (A.C.W.), and diagnosis of AD, if present, was made according to NINCDS criteria. Diagnosis of AD required a history of deterioration in memory, cognitive function, and adaptive function; exclusion by clinical examination of focal neurological or systemic conditions that could account for the deterioration; and objective evidence of cognitive impairment on a standardized cognitive screening test (a score≤23 on the Mini-Mental State Examination8).
APOE genotyping was performed by a modification of the method of Wenham et al.9 Genotype scorings were made independently by two researchers and compared.
RESULTS
In the Forty Families, there were 121 individuals over the age of 65, and 1 was affected by AD (APOE genotype was 3/3). Of another 76 individuals over age 60 in the adjacent church districts, 4 were affected. Thus, 5 of 197 individuals had AD. This is well below the 11.82 expected (95% confidence interval by Poisson distribution: 0.62–9.38) if a prevalence of 6% in the general population is assumed.
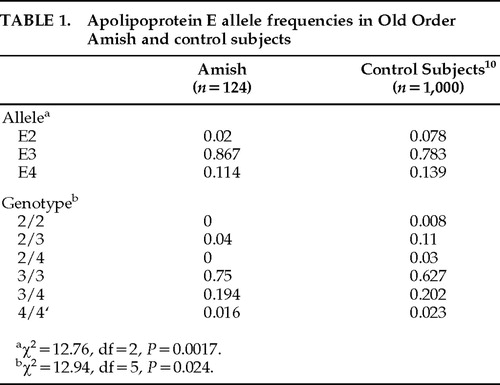
APOE allele frequencies are shown in Table 1 for the Old Order Amish individuals we studied and for general population control subjects as described by Menzel et al.10 Compared with general population control subjects, the Amish had significantly different frequencies of APOE alleles (χ2=12.76, df=2, P=0.0017, 2-sided) and significantly different genotype frequencies (χ2=12.94, df=5, P=0.024). The E4 allele frequencies were not significantly reduced in the Amish compared with control subjects (χ2=1.17, P=0.28). The E2 allele frequencies were significantly reduced (χ2=10.71, df=1, P=0.001) in the Amish.
DISCUSSION
In the individuals we studied from the Old Order Amish of Lancaster County, we found a decreased prevalence of AD, a lower frequency of the E2 allele, and a normal E4 allele frequency compared with the general population. We do not confirm the results of Pericak-Vance et al.7 of a lower frequency of E4 in the Amish. Although we do confirm their suggestion of a reduced prevalence of AD in the Amish, our data do not support their hypothesis that this may be partially explained by a reduced frequency of the E4 allele. The decreased frequency of the E2 allele rules out the possibility that the decreased prevalence of AD is caused by more individuals being protected by the E2 allele.
The reason for the reduced prevalence of AD is not clear. Environmental influences or other differences in genetic background could be involved: the Amish way of life differs from that of the general population in many aspects, including in family structure, education, diet, health care, type of work experience, social interactions, and use of technology. Communities are predominantly rural.
The finding of different APOE allele frequencies in the Amish compared with the general population, and between two separated Amish populations, is not surprising. The Old Order Amish are a genetically isolated population with a well-defined European ancestry. The population is closed, since the Amish do not proselytize and marrying outside the faith is forbidden. The Old Order Amish of Lancaster County are a different community from those studied by Pericak-Vance et al.,7 although the Indiana community was founded by migration from the original Pennsylvania community.11 The Indiana Amish and Lancaster County Amish have been previously proposed to be partially genetically distinct.12
There are several limitations to our study. The number of individuals screened was relatively small, and APOE genotypes were obtained on a selected group of individuals, not a whole population. The identification of affected individuals required sufficient cognitive deterioration to have been noticeable to the Amish research worker; more subtle impairments might have been detectable in other individuals by use of standardized psychological tests, but the demonstration of deterioration would have required a longitudinal study.
The large and stable family structures of the Amish make them attractive for genetic studies, and they are a valuable resource to search for families with multiple affected individuals. We had marked difficulty finding such families even among the highly inbred13 Lancaster County Amish, as might be expected from the lower prevalence of AD. Similarly, although there are 7 affected individuals in the extended pedigree of Pericak-Vance et al.,7 only 2 of these individuals are from the same sibship, and the next closest relationship is at the level of third-degree relatives. It appears that it might be more fruitful to search for families with multiple individuals with AD in genetically isolated populations other than the Amish.
It is interesting to note that the Amish also have a low rate of coronary artery disease compared with the general population; deaths in Amish males from ischemic heart disease are at approximately 60% of the expected frequency.11 An association between APOE E2, APOE E4, and ischemic heart disease has been found in previous studies14; however, it is not clear whether reduced E2 and E4 allele frequencies in the Amish contribute to their reduced death rate.
ACKNOWLEDGMENTS
The authors are grateful for DNA from Amish families provided by Drs. Wilma Bias and Barbara Schmeckpeper.
 |
1. Goate A, Chartier-Harlin M-C, Mullan M, et al: Segregation of a missense mutation of the amyloid precursor protein gene with familial Alzheimer's disease. Nature 1991; 349:704–706Crossref, Medline, Google Scholar
2. Sherrington R, Rogaev EI, Liang Y, et al: Cloning of a novel gene bearing missense mutations in early familial Alzheimer disease. Nature 1995; 375:754–760Crossref, Medline, Google Scholar
3. Levy-Lehad E, Wijsman EM, Nemens E, et al: A familial Alzheimer's disease locus on chromosome 1. Science 1995; 269:970–973Crossref, Medline, Google Scholar
4. Saunders AM, Strittmatter WJ, Schmechel D, et al: Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology 1993; 43:1467–1472Crossref, Medline, Google Scholar
5. Corder EH, Saunders AM, Risch NJ, et al: Apolipoprotein E type 2 allele decreases the risk of late onset Alzheimer disease. Nature Genet 1994; 7:180–184Crossref, Medline, Google Scholar
6. Schellenberg GD, Bird TD, Wijsman EM, et al: Absence of linkage of chromosome 21q21 markers to familial Alzheimer's disease. Science 1988; 241:1507–1510Crossref, Medline, Google Scholar
7. Pericak-Vance MA, Johnson CC, Rimmler JB, et al: Alzheimer's disease and apolipoprotein E-4 allele in an Amish population. Ann Neurol 1996; 39:700–704Crossref, Medline, Google Scholar
8. Folstein MF, Folstein SE, McHugh PR: “Mini-Mental State”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975; 12:189–198Crossref, Medline, Google Scholar
9. Wenham PR, Price WH, Blundell G: Apolipoprotein E genotyping by one-stage PCR. Lancet 1991; 337:1158–1159Crossref, Medline, Google Scholar
10. Menzel H-J, Kladetzky R-G, Assmann G: Apolipoprotein E polymorphism and coronary artery disease. Arteriosclerosis 1983; 3:310–315Crossref, Medline, Google Scholar
11. Hamman RF: Patterns of mortality in the Old Order Amish. DPH dissertation, Johns Hopkins University, Baltimore, MD, 1979Google Scholar
12. McKusick VA, Hostetler JA, Egeland JA: Genetic studies of the Amish. Bulletin of the Johns Hopkins Hospital 1964; 115:203–222Medline, Google Scholar
13. McKusick VA: Medical Genetic Studies of the Amish: Selected Papers. Baltimore, MD, Johns Hopkins University Press, 1978Google Scholar
14. Eto M, Watanabe K, Makino I: Increased frequencies of apolipoprotein ε2 and ε4 alleles in patients with ischemic heart disease. Clin Genet 1989; 36:183–188Crossref, Medline, Google Scholar